Protein fluorescence localization detecting system and method for fast detecting protein interaction and application thereof
A protein interaction and positioning detection technology, applied in the fields of biotechnology and genetic engineering, can solve the problems of prone to false positives, many false positives, and can not be guaranteed, and achieve the effect of eliminating false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
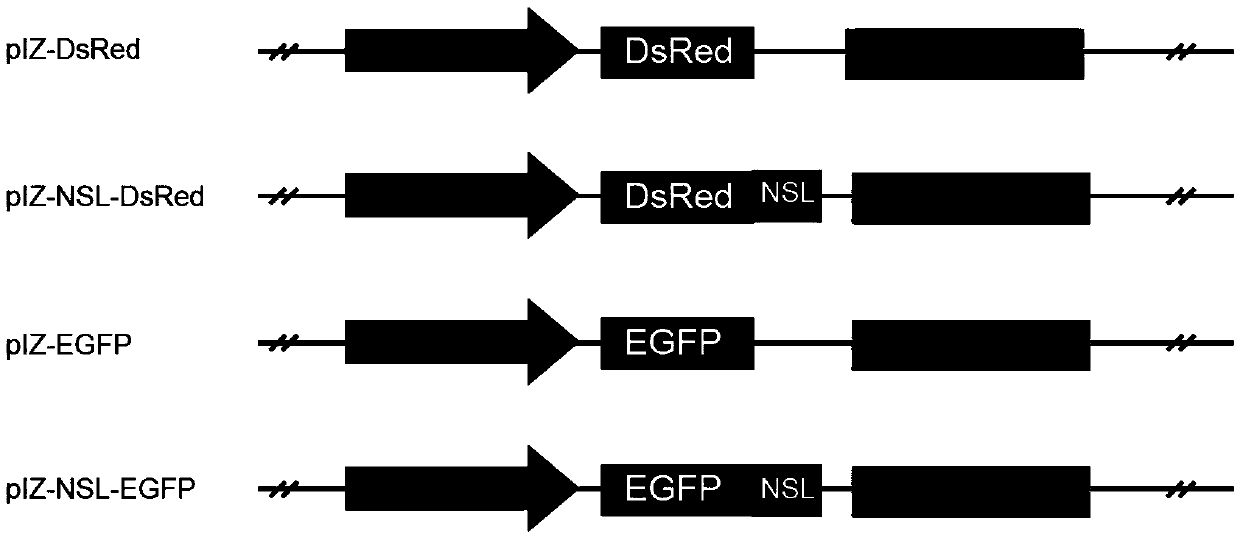
[0035] Example 1. Protein fluorescence localization detection carrier construction
[0036] Log in to the NCBI database and download the currently known nucleotide sequences expressing DsRed and EGFP proteins, the sequences of which are shown in SEQIDNo.1 and SEQIDNo.2 respectively, according to the multiple cloning site of the pIZ-V5 / His (Invitrogen Company) vector Design corresponding primers with nuclear localization signal sequence and without nuclear localization signal sequence, and then send them to BGI for synthesis. The primer sequences are as follows:
[0037] EGFP-F:5'-cccaagcttatggtgagcaagggcgaggagct-3', (SEQ ID No.10);
[0038] EGFP-R:5'-ggggtacccttgtacagctcgtccatgcc-3', (SEQ ID No.11);
[0039] NSL-EGFP-F:5'-cccaagcttccgaagaagaagcgaaaggtggaagacccgggaacgatggtgagcaagggcgaggagct-3', (SEQ ID No. 12);
[0040] DsRed-F:5'-cccaagcttatggcctcctccgagaacgtcat-3', (SEQ ID No.13);
[0041] DsRed-R: 5'-ggggtacccaggaacaggtggtggcgg-3', (SEQ ID No. 14);
[0042] NSL-DsRed-F: ...
Embodiment 2
[0046] Example 2, Verification of Protein Interaction Identification by Protein Fluorescence Localization Detection System
[0047] The protein fluorescent localization carrier constructed in Example 1 can be localized to the corresponding position. In order to verify that this system can observe whether there is an interaction between the two proteins through the change of fluorescent localization, first analyze that LEF-11 has a nuclear localization signal, and select pIZ- EGFP and pIZ-DsRed were used as target vectors, and a full-length fusion vector pIZ-DsRed-LEF11 / pIZ-EGFP-LEF11 of LEF-11 (sequence shown in SEQIDNo.4) full-length fusion of DsRed and EGFP was constructed, and a mutant LEF-11 was constructed at the same time The key amino acid mutant sequence of the nuclear localization signal at position 100 is shown in SEQ ID No.5; the vector pIZ-LEF11(K100L)-DsRed fused with DsRed, and the control vector is pIZ-EGFP. The vector construction method is the same as that in ...
Embodiment 3
[0051] Example 3. Application of protein fluorescence localization detection system in the study of LEF-11 protein interaction with nuclear localization signal
[0052] In order to study the practicability of the protein fluorescence localization detection system described in Example 1 in identifying protein interactions, the N-terminus of LEF-11 containing only one oligomerization domain (amino acid 42-61 region) was truncated and mutated, And a fluorescent protein fusion expression vector was constructed, see for details Image 6 Schematic diagram of fluorescent protein construction of LEF-11 N-terminal truncation mutant vector. The specific construction method of the vector is the same as that in Example 1, and all the vectors have been verified by enzyme digestion and sequencing.
[0053] The constructed fluorescent protein fusion expression vector pIZ-DsRed-LEF11(aa2-61), pIZ-DsRed-LEF11(aa12-61), pIZ-DsRed-LEF11(aa22-61), pIZ-DsRed-LEF11(aa32-61 ), pIZ-DsRed-LEF11(aa42...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com