MicroRNA SDA detecting method based on AgNCs/HpDNA probes
A detection method, 45s-1min technology, applied in DNA/RNA fragments, recombinant DNA technology, microbial determination/inspection, etc., can solve the problem of difficulty in ensuring specificity, and achieve the effect of shortening reaction time and saving reaction materials
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
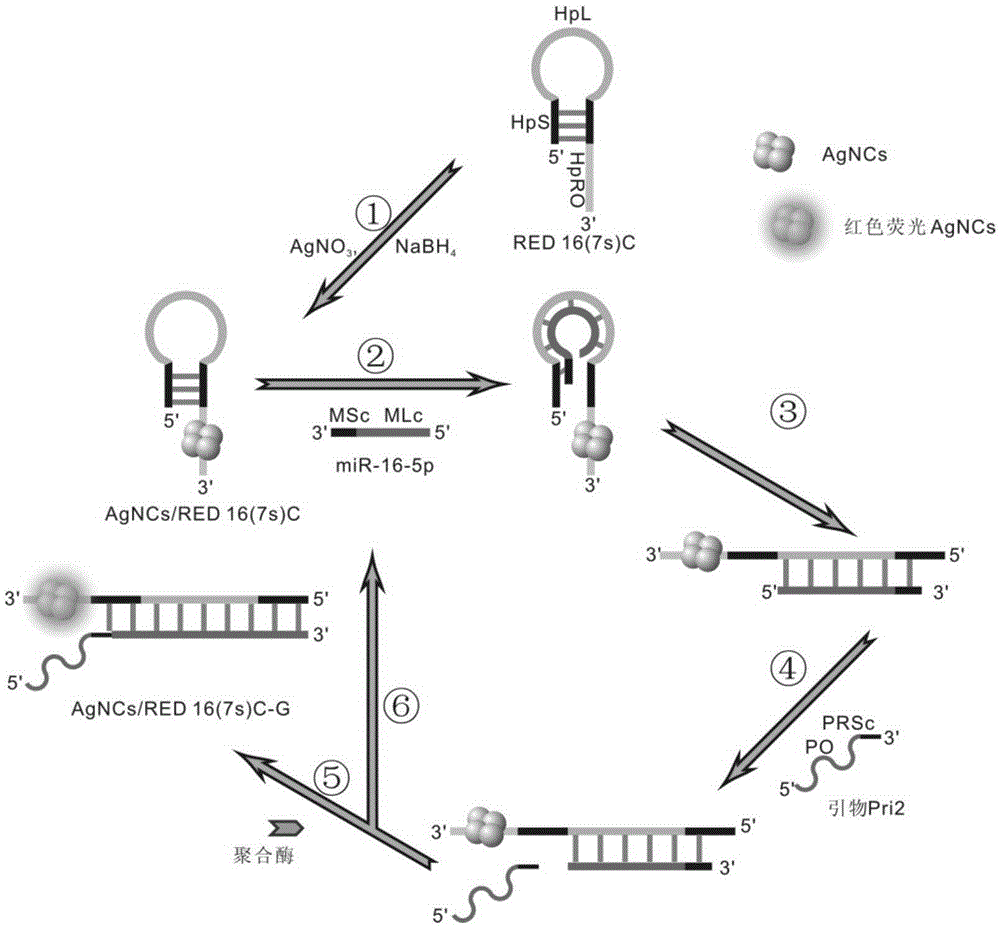
[0051] Embodiment 1, detection scheme feasibility verification
[0052] (1) AgNCs / HpDNAs probe synthesis
[0053] The synthesis was carried out according to the method in Yeh. HpDNAs (RED16(7s)C), AgNO 3 , NaBH 4 The starting concentrations were 100 μM, 1 mM and 1 mM, respectively. The stock concentration of phosphate buffer was 200mM (Pi, pH8.0). Equimolar AgNO 3 and NaBH 4 According to 1RED16(7s)C:25AgNO 3 :25NaBH 4 The proportion of the three was added to HpDNA, so that the final concentrations of the three were 15μM, 375μM and 375μM (Pi, 20mM, pH8.0). Among them, NaBH 4 It needs to be freshly prepared, and finally quickly added to the Ag+ / HpDNA mixture within 30s, and then shake vigorously for 45s~1min. The resulting solution was placed at room temperature in a dark environment for 18 hours to obtain stable AgNCs / HpDNAs probes.
[0054] (2) Verification of fluorescence enhancement effect of G-rich sequence hybridization
[0055] To the obtained probe AgNCs / RE...
Embodiment 2
[0058] Embodiment 2, single miRNA detection
[0059] (1) AgNCs / RED16(7s)C probe synthesis
[0060] Prepare the AgNCs / RED16(7s)C probe as described in (1), but the final concentration of RED16(7s)C is 5 μM, AgNO 3 and NaBH 4 The amount added is still according to 1RED16(7s)C:25AgNO 3 :25NaBH 4 conduct.
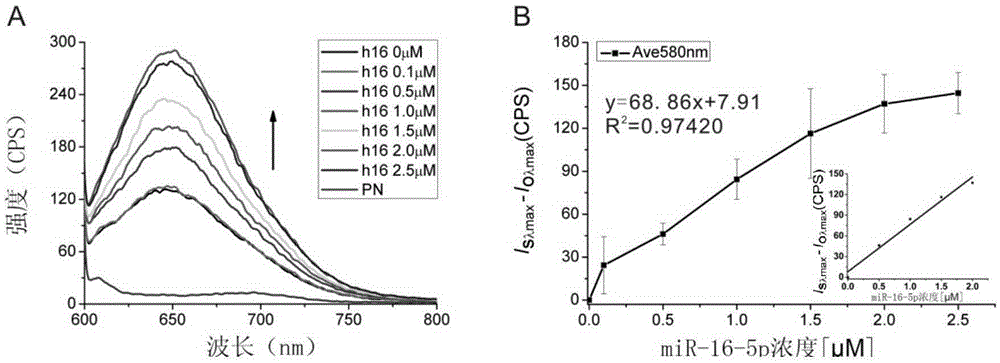
[0061] (2) Single miRNA detection
[0062] Each reaction tube contains 50μL SDA reaction solution, which contains the following components: 1×Nb2.1 homemade buffer (buffer pH 7.925°C) (50mMNaAc, 10mMTris-HAc, 10mMMg(Ac)2 and 100 μg / mL BSA) 200 μM dNTPs, 10 UBsu polymerase (without DTT), AgNCs / RED16(7s)C (2.5 μM HpDNA), different concentrations of target miRNA (0.05-2.5 μM miR-16-5p), and 2.5 μM primer Pri2. The obtained reaction solution was incubated at 55°C for 55min, and then stored in a dark environment at 4°C, then the fluorescence detection could be performed on a fluorescence spectrophotometer, and the experiment was repeated 3 times.
[0063] (3) Results
[006...
Embodiment 3
[0066] Embodiment 3, single base mismatch nucleic acid detection
[0067] (1) Single base mismatch nucleic acid detection
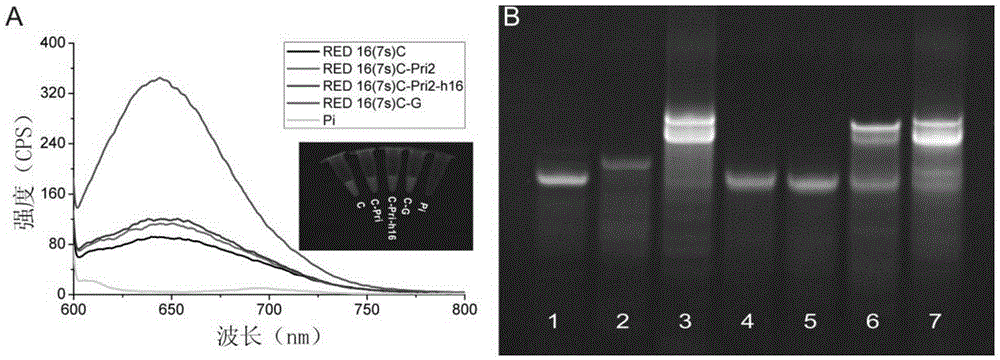
[0068] According to the steps described in (1, 2) in Example 2, only the nucleic acid to be tested is changed to 0.5 μM mismatched nucleic acid, that is, based on the AgNCs / RED16 (7s) C probe to detect miR-19b-3p, h16DMⅠ (sequence SEQ ID NO :5) and h16DMII (SEQ ID NO: 6).
[0069] (2) Results
[0070] Mismatch nucleic acid detection results such as Figure 4 shown. It can be seen that in the detection based on the AgNCs / RED16(7s)C probe, only miR-16-5p (λex=580nm) showed the highest fluorescence enhancement signal, and the fluorescence enhancement of other samples at this excitation wavelength was lower than Weak, especially, h16DMⅠ showed fluorescence quenching under excitation of 580nm wavelength. In summary, the detection based on the AgNCs / RED16(7s)C probe showed good specificity.
[0071] The results of gel electrophoresis showed that the mis...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com