A SNP Molecular Marker of Jute Anthracnose Resistance Gene Segment
A technology of molecular markers and resistance genes, which is applied in the direction of recombinant DNA technology, microbial detection/testing, DNA/RNA fragments, etc., can solve the problems of reducing jute yield and fiber quality, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
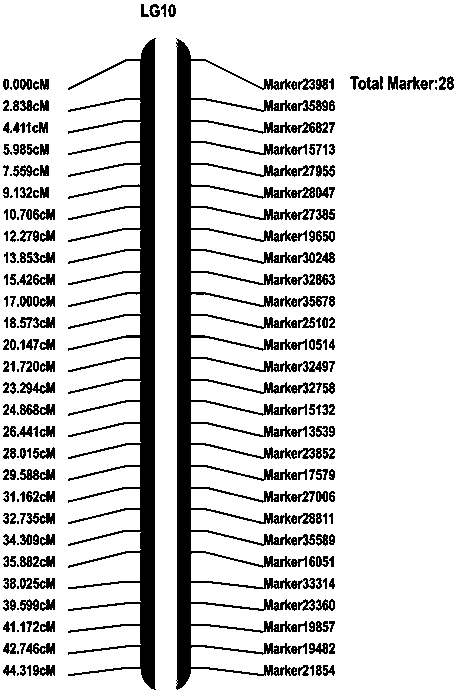
[0076] Example 1: 26 SNP molecular markers linked to anthrax resistance genes were developed, such as figure 2 shown.
Embodiment 2
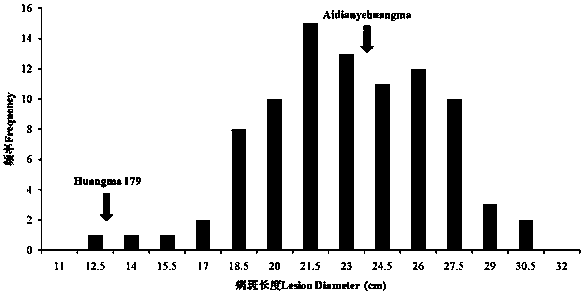
[0077] Example 2: Using the SNP molecular markers developed above for jute resistance germplasm screening. The test materials were 144 jute lines. 18 disease-resistant strains numbered 98, 45, 76, 122, 124, 20, 101, and 27 were screened out, accounting for 12.5% of the total. It shows that there are obvious differences in the resistance to anthracnose among different jute strains. In order to further confirm the accuracy of field inoculation, 7 strains with different resistances were selected for inoculation identification in the greenhouse. Investigating its incidence, it was found that there was little difference from field inoculation.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com