Function of purple acid phosphatase GmPAP33 gene for promoting reuse of phosphorus in soybean mycorrhiza
An acid phosphatase and gene technology, applied in the field of genetic engineering, can solve problems such as unclear specific functions, and achieve the effect of improving the ability of efficient phosphorus utilization and improving the phosphorus efficiency of plants
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
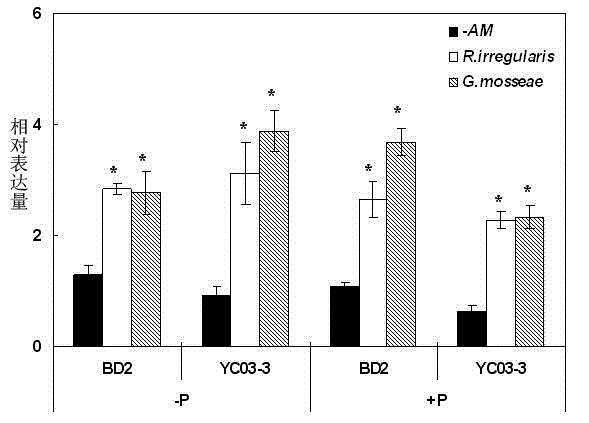
[0032] GmPAP33 Gene expression pattern analysis: In the published soybean genome database, through homologous comparison, it was predicted that there are 35 members of soybean purple acid phosphatase, which were determined by quantitative PCR technology. GmPAP33 Genes are strongly expressed genes in mycorrhizae. GmPAP33 The length of the open reading frame of the gene is 1290 bp, encoding a protein of 429 amino acids. Studies have shown that this gene is closely related to soybean arbuscular mycorrhizal fungal infection, and its expression pattern was first analyzed.
[0033]Two soybean varieties were used, phosphorus low-efficiency local 2 (BD2) and phosphorus-high efficiency Yuechun 03-3 (YC03-3). Set 3 AMF inoculation treatments as non-inoculation (-AM) and inoculation with AM fungi Rhizophagus irregularis or AM fungi Glomus mosseae . Set up two phosphorus treatments, 5 μM KH 2 PO 4 As low phosphorus (-P) and 250 μM KH 2 PO 4 K as high phosphorus (+P), low phosph...
Embodiment 2
[0043] 1, GmPAP33 Gene cloning: using soybean flowering leaf cDNA as template, using upstream specific primer F1: 5'- GAGCTC ATGGGGATGCGAATG -3' (SEQ ID NO:5) and downstream specific primer R2: 5'- TCTAGA CTATTTACAGGAAGGAAT -3' (SEQ ID NO: 6) amplification GmPAP33 The ORF full-length sequence of the gene is 1290bp, and sequenced and compared, successfully obtained GmPAP33 A coding sequence that encodes a protein of 429 amino acids.
[0044] 2, GmPAP33 Gene promoter cloning, vector construction and tissue expression localization analysis: Promoter analysis Construction of expression vector: According to conventional methods, extract genome DNA of soybean YC03-3 genotype at the flowering stage, use soybean leaf genome DNA as template, and use upstream Specific primer F2: 5'-CCG GAATTC GAAAGCGTTGGGCTTAAAC-3' (SEQ ID NO: 7) and downstream specific primer R2: 5'-CTAG TCTAGA Amplification of TGTTTTCAAATTTCGGTGC-3' (SEQ ID NO:8) GmPAP33 The 2019 bp fragment of the pr...
Embodiment 3
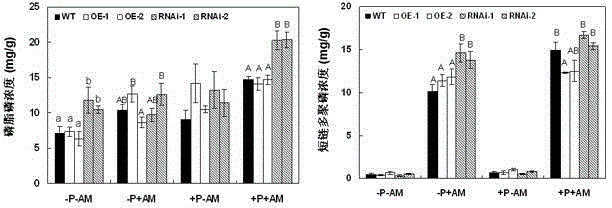
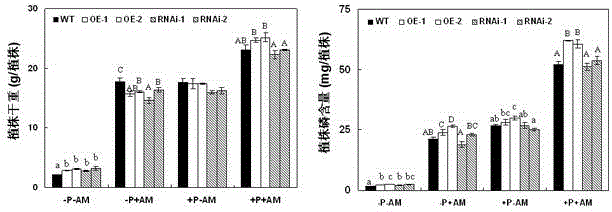
[0048] 1. Overexpression GmPAP33 The construction of the vector: using the cDNA of the leaf part of the flowering stage of the soybean YC03-3 genotype as a template, using the upstream specific primer F1: 5'- GAGCTC ATGGGGATGCGAATG-3' (SEQ ID NO:5) and downstream specific primer R1: 5'- TCTAGA CTATTTACAGGAAGGAAT-3' (SEQ ID NO: 6) amplification GmPAP33 The full-length sequence of ORF is 1209 bp. After the PCR recovery fragment is sequenced correctly, pass Sac I and Xba After performing double enzyme digestion on the fragment and the target vector, the GmPAP33 The gene was connected to the destination vector pTF101.1. Transform Escherichia coli DH10B, transform Agrobacterium tumefaciens EHA101 after the sequence is correct, and use for Agrobacterium tumefaciens-mediated transformation of whole soybean cotyledon nodes.
[0049] 2. Interference expression GmPAP33 Construction of the vector: using the cDNA of the leaf at the flowering stage of the soybean YC03-3 genotyp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com