Method and set for detecting alcohol metabolizing genes
A gene and genotype technology is applied in the field of detecting alcohol dehydrogenase and acetaldehyde dehydrogenase, and can solve the problems of complicated genotype polytype detection steps, difficulty in a large number of screening tests, and long time consumption.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0047] The primers of the present invention can be prepared using methods known in the art, including, but not limited to, cloning and designing appropriate sequences and direct chemical synthesis.
[0048] Chemical synthesis methods that can be used to prepare primers of the invention include, but are not limited to, the phosphotriester method described in Narang et al., Methods in Enzymology, 68:90 (1979), the method described in Brown et al., Methods in Enzymology, 68:109 (1979) and the solid phase synthesis described in US 4,458,066; automated oligonucleotide synthesizers may also be used. In addition, primers can be labeled, if desired, using techniques known in the art.
[0049] The aforementioned oligonucleotide primer set of the present invention can be used for amplifying methods (such as polymerase chain reaction (polymerase chain reaction, PCR) and modification methods thereof) to identify and detect the genotype (ADH1B) of the single nucleoside polymorphism R49H of...
example 1
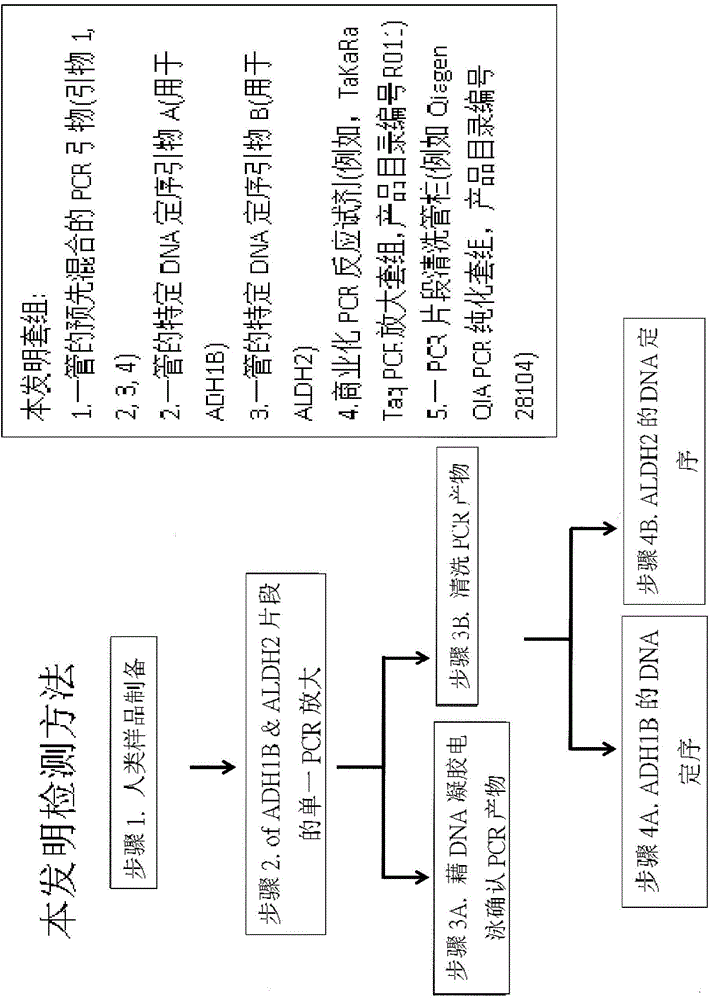
[0069] Genotype Detection of Example 1 Human ADH1B and ALDH2
[0070] Test samples such as blood (200 μl), saliva (2 ml) or cells are obtained from human subjects. Genomic DNA was purified and extracted with a DNA purification kit (Qiagen DNeasy Blood & Tissue Kit. Cat. No. 69504 or 69581)). Mix the primers in the following table in 1 tube, and use TaKaRa Taq PCR Amplification Kit to perform PCR reaction to obtain ADH1B and ALDH2 fragments. The PCR cycle was: 94°C, 1 minute; (94°C, 10 seconds; 62°C, 20 seconds; 68°C, 30 seconds); performed 35 cycles; 72°C, 10 minutes; and maintained at 4°C.
[0071]
[0072] Then use gel electrophoresis to confirm that the amplified product contains the target PCR fragment (the fragment of ADH1B (R49H) and ALDH2 (E487K) single nucleoside polymorphism) (please refer to Figure 4 ), and then purify the target PCR fragment with Qiagen QIA PCR purification kit.
[0073] The aforementioned target PCR fragments, PCR primer sets and fragment si...
example 2
[0076] Example 2 Relationship between genotypes of ADH1B (R49H) and ALDH2 (E487K) single nucleoside polymorphisms and cancer risk
[0077] According to the literature published in PLoS Medicine, March 2009, Vol.6, Issue 3, pp.0258-0263 and Yokoyama et al. Keio J Med 2010; 59(4): 115-130, the present invention re-counts and organizes ADH1B and The relationship between ALDH2 genotype, flushing after alcohol consumption, and cancer risk is shown in the table below:
[0078]
[0079]
[0080] Based on the above table, it can be seen that people with ADH1B*1 / *1 and ALDH2*1 / *2 genotypes have a higher risk of cancer. Therefore, the methods and kits of the present invention can be used to screen individuals at higher risk of developing cancer after consuming alcohol.
[0081]
[0082]
[0083]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com