A method of detecting quantum dot-protein binding kinetics
A quantum dot and kinetic technology, applied in the direction of electrochemical variables of materials, etc., can solve the problems of difficult separation of quantum dot-protein complexes, low resolution, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Detection of dBSA and QDs Kinetics in Capillary
[0021] 1. Fat-soluble QDs are converted into water-soluble QDs by GSH
[0022] Mix 18 mg GSH, 5 mg KOH, and 250 μL methanol, take 40 μL of the mixed solution and add it to 200 μL fat-soluble quantum dots, and shake for 30 minutes. After shaking, 200 μL of 1 mM NaOH was added, and the fat-soluble quantum dots were transferred to the water phase. The quantum dots in the upper layer were taken out, precipitated by adding 1 mL of methanol and 30 μL of NaCl solution (30 mg / mL), and dissolved in a pH 7.4 boric acid buffer. The precipitation was repeated twice, and finally dissolved in 200 μL pH7.4 boric acid buffer. The concentration of QDs does not change after switching.
[0023] 2. Preparation of dBSA solution
[0024] dBSA solution through NaBH 4 Prepared by reacting with BSA. The main steps are as follows: 0.33g BSA was dissolved in 100mL pH 7.4 borate buffer, then 0.008g NaBH was added while stirring 4 , stirred a...
Embodiment 2
[0030] Extracapillary detection of dBSA and QDs kinetics
[0031] Steps 1 and 2 are the same as in Example 1.
[0032] 3. QDs coupled with dBSA
[0033] Different ratios of dBSA and QDs were self-assembled outside the capillary for 60 s, and then injected for 20 s and detected by electrophoresis.
[0034] 4. Fluorescence capillary electrophoresis analysis and detection
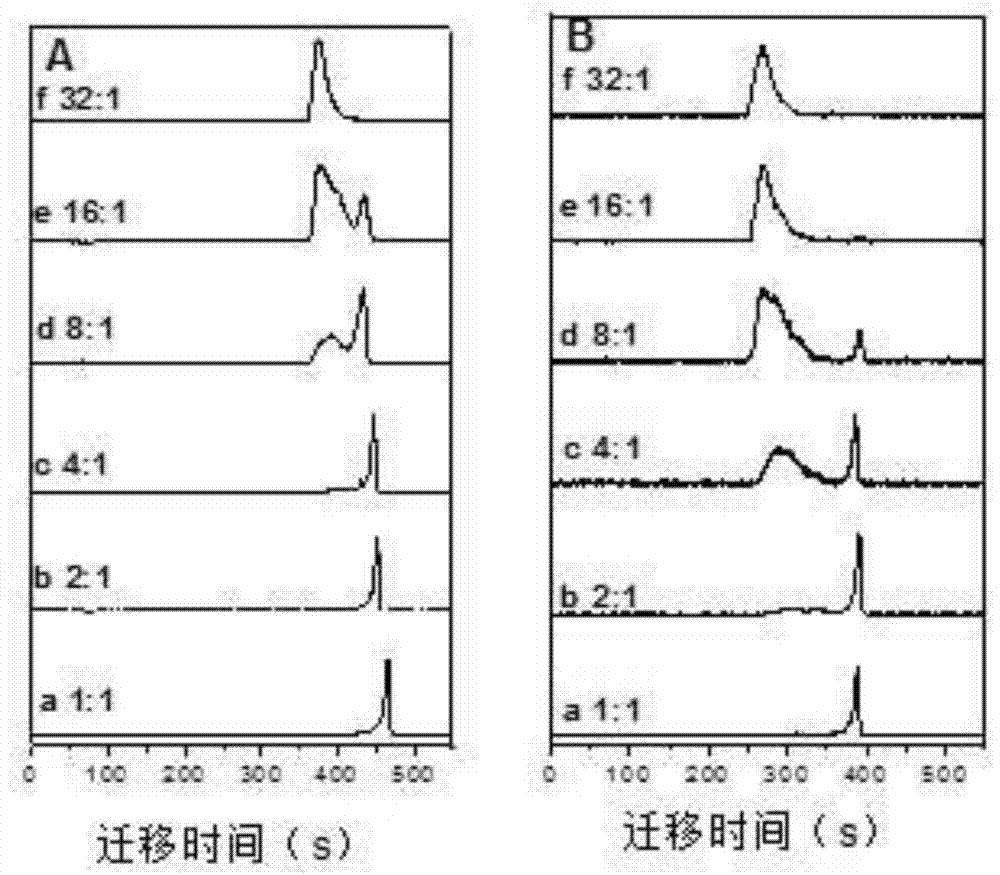
[0035] When we detected outside the capillary, with the increase of dBSA concentration, the electrophoretic peak of QDs gradually decreased, and the electrophoretic peak of the complex gradually increased, and the reaction was complete at 16:1 ( figure 1 B). According to S complex / S total and dBSA concentration, QDs and dBSA kinetic curves were obtained by Hill equation fitting ( figure 2 , curve b). K is calculated by the Hill equation D 2.8×10 -6 M, n is 2.8, R 2 is 0.994.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com