A pathogenic mutation of hereditary cone dystrophy and its detection reagent
A cone cell and malnutrition technology, applied in the field of biomedicine, can solve the problems of not being able to locate the disease-causing site, analyzing small families and sporadic cases, and screening out disease-causing genes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] A second-generation family with cone dystrophy (CD) was detected for mutations in the LCA5 gene.
[0046] experimental method:
[0047] 1. Collection of clinical resources of the family and establishment of a genetic resource bank:
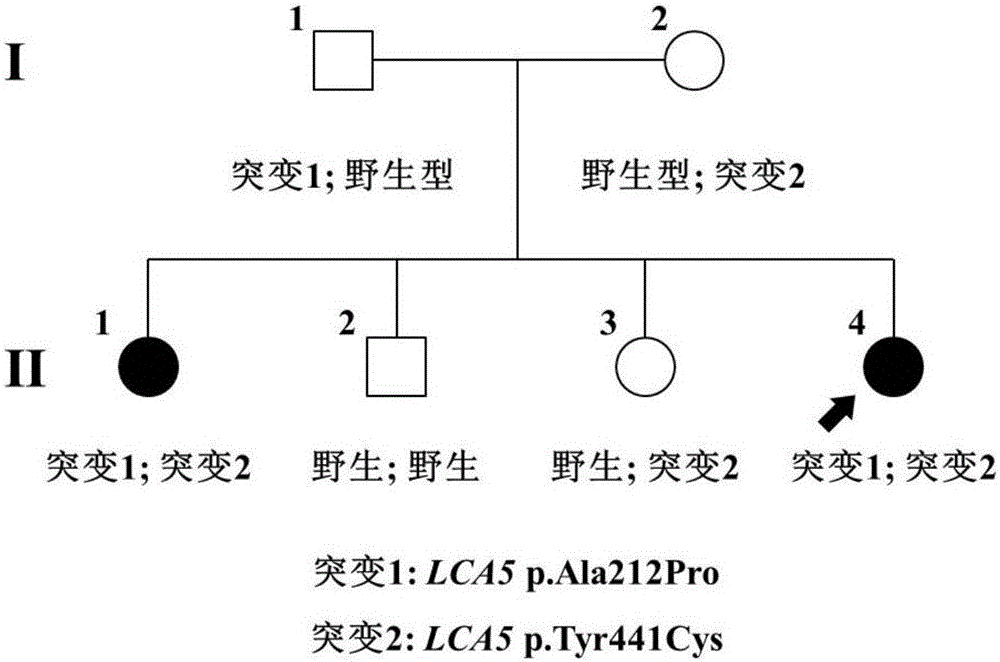
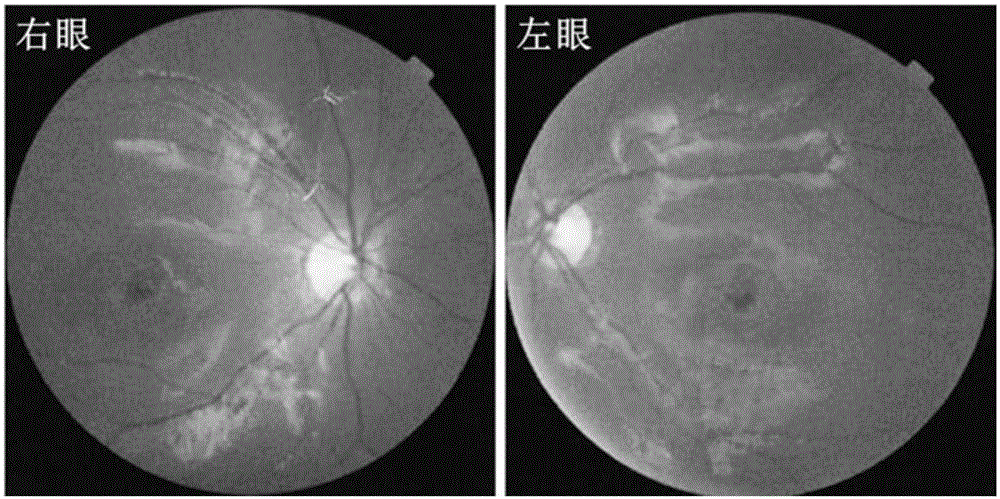
[0048] The clinical data and blood samples of each member of the family were collected, see the family diagram figure 1 . Clinical data mainly include personal medical history, family history, best corrected visual acuities (BCVAs), slit lamp examination, fundus photography, color vision examination, visual field examination (Humphrey perimetry), visual evoked potential detection (visual-evokedpotentials); VEP), full field electrophysiology (electroretinography; ERG), fundus fluorescein angiography (FFA) and optical coherence tomography (optical coherence tomography; OCT), etc. The blood genomic DNA of each family member was extracted with a blood genomic DNA extraction kit (Qiagen, Hilden, Germany).
[0049] 2. Discover the pathogenic ...
Embodiment 2
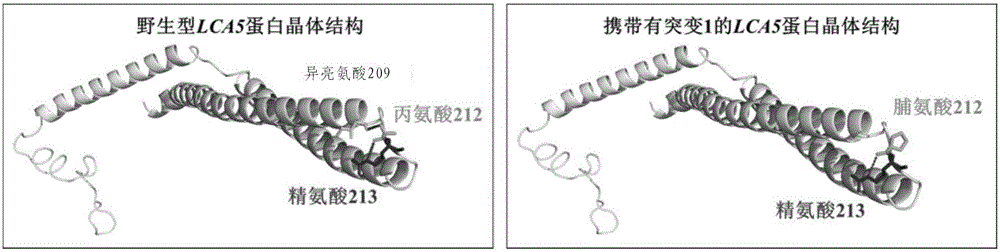
[0082] Functional studies were carried out on the pathogenic genes detected in Example 1. Here, the new mutation p.[Ala212Pro];[Tyr441Cys] detected in the LCA5 gene was taken as an example.
[0083] experimental method:
[0084] 1. Conservative analysis:
[0085] Using NCBI HomoloGene database ( http: / / www.ncbi.nlm.nih.gov / homologene ) Conservative evaluation and prediction of the screened mutations in multiple species.
[0086] 2. Predict the pathogenicity of mutations based on SIFT and PolyPhen values:
[0087] Using two mainstream online prediction software: PolyPhen-2 (Polymorphism Phenotyping, version2; http: / / genetics.bwh.harvard.edu / pph2 / ) and SIFT Human Protein DB ( http: / / sift.bii.a-star.edu.sg / ), to predict the impact of missense mutations and nonsense mutations on protein levels, thereby predicting the pathogenicity of mutations.
[0088] 3. Research on protein crystal structure changes:
[0089] SWISS MODEL (http: / / swissmodel.expasy.org / ) prediction s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com