Multiple testing methods for gene mapping
A technology of gene positioning and testing methods, applied in the field of bioinformatics, to achieve the effect of improving precision and accuracy, improving utilization rate, and shortening cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] Embodiments of the present invention are described in further detail below in conjunction with accompanying drawings:
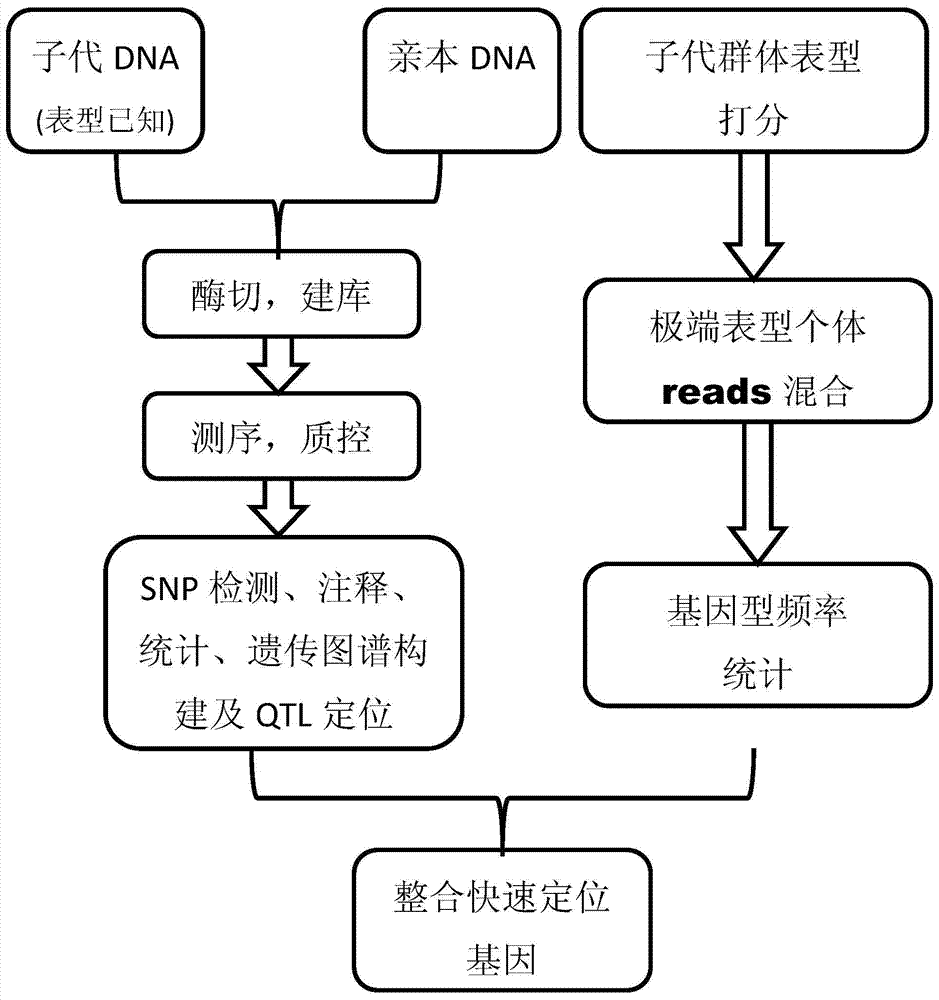
[0028] The invention discloses a multiple testing method for gene positioning, which is characterized in that it comprises:
[0029] Step 1: Sequence the parental DNA samples and offspring DNA samples using second-generation sequencing technology to obtain high-precision short-segment sequences;
[0030] Step 2: using SNP analysis software to compare or cluster the obtained high-precision short fragment sequences with reference sequences to obtain accurate SNP information for each sample;
[0031] Step 3: convert such SNP information into population SNP, further convert into standard molecular standard format, use mapping software to construct genetic map, and perform QTL analysis on the basis of genetic map to obtain QTL locus information;
[0032] Step 4: Identify the phenotypic information of the progeny DNA samples, and accurately count the distri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com