Method for constructing gene 2a type hepatitis c virus clinical isolates cell culture model
A hepatitis C virus, hepatitis C virus technology, applied in the fields of biotechnology and virology, can solve the problem of limited virus strains
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
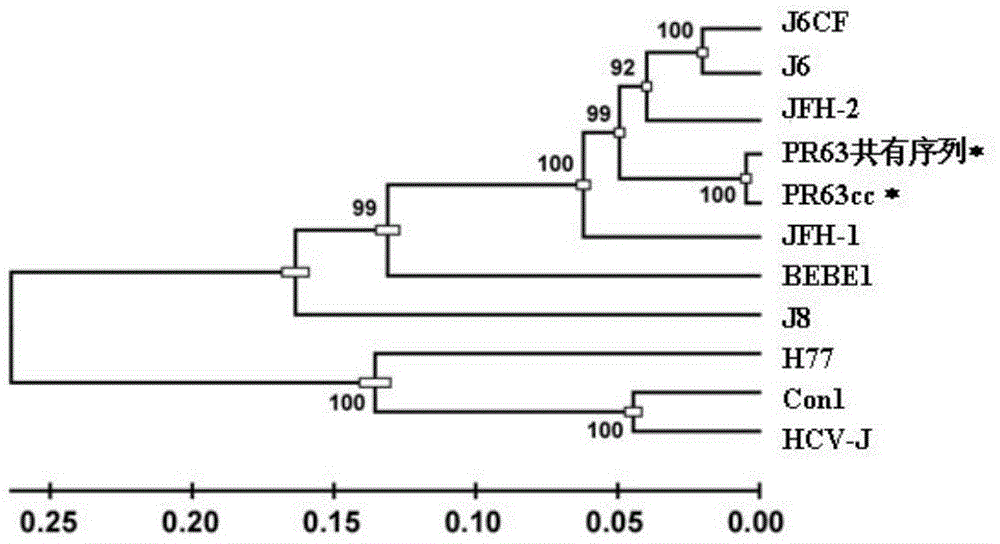
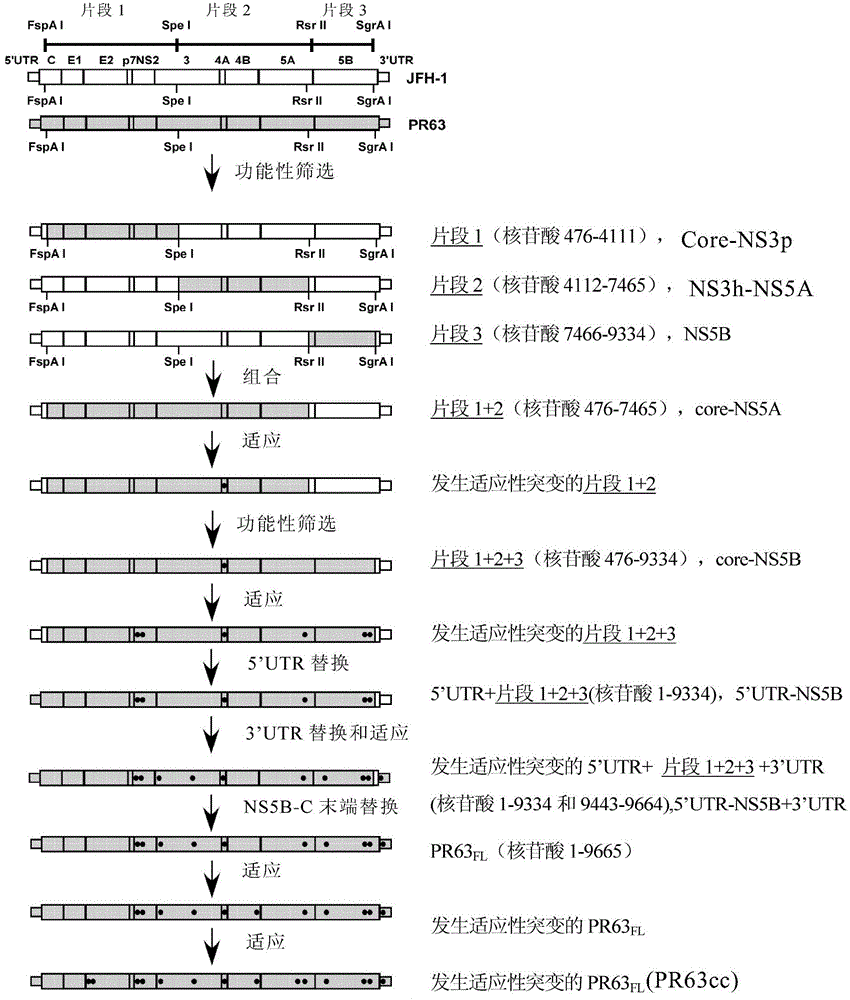
[0072] Example 2. Core protein-NS3 protease region, NS3 helicase-NS5A region and NS5B region of PR63 are active in in vitro culture
[0073]Except for JFH-1, other HCV clones were inactive in cell culture: the RNA of wild-type MA, J6CF, and DH8CF could not replicate and produce infectious particles in cultured cells (Li, Y. et al., 2012, Proceedings of the National Academy of Sciences of the United States of America 109:E1101-1110; Murayama, A., T. et al., 2012, Journal of virology 86:2143-2152; Ramirez, S. et al., 2013, Hepatology (Baltimore, Md)). Therefore, the present inventors did not start from PR63 to construct its consensus sequence cDNA clone. On the contrary, they first used JFH-1 as the backbone to target the core protein-NS3 protease region containing the source of the PR63 sequence (the upstream of the SpeI enzyme cleavage point in the NS3 coding sequence, NS3p), NS3 helicase (downstream of the SpeI cut point in the NS3 coding sequence, NS3h)-NS5A region, NS5B reg...
Embodiment 3
[0078] Example 3. Combining functional regions of PR63 and confirming mutations that can promote virus production
[0079] The present inventors tried to combine PR63-derived FspA I-Spe I segment, Spe I-Rsr II segment and Rsr II-SgrA I segment to construct a nearly full-length infectious clone of PR63. However, these three segments were spliced together to replace the corresponding region in the JFH-1 vector, resulting in clone PR63 C-NS5B / JFH-1 (selected from: FspA I-Spe I segment from S10 + Spe I-Rsr II segment from A6 + Rsr II-SgrA I segment from 5B13 (S10-A6-5B13) (from FspA I-SgrA The sequence between I is the clone derived from PR63, and the rest is still the sequence derived from JFH-1); FspA I-Spe I segment from S10 + Spe I-Rsr II segment from A6 + Rsr II-SgrA from 5B21 I segment (S10-A6-5B21) or FspA I-Spe I segment from S10 + Spe I-Rsr II segment from A6 + Rsr II-SgrA I segment from 5B23 (S10-A6-5B23)) after electroporation No HCV-positive cells were found in th...
Embodiment 4
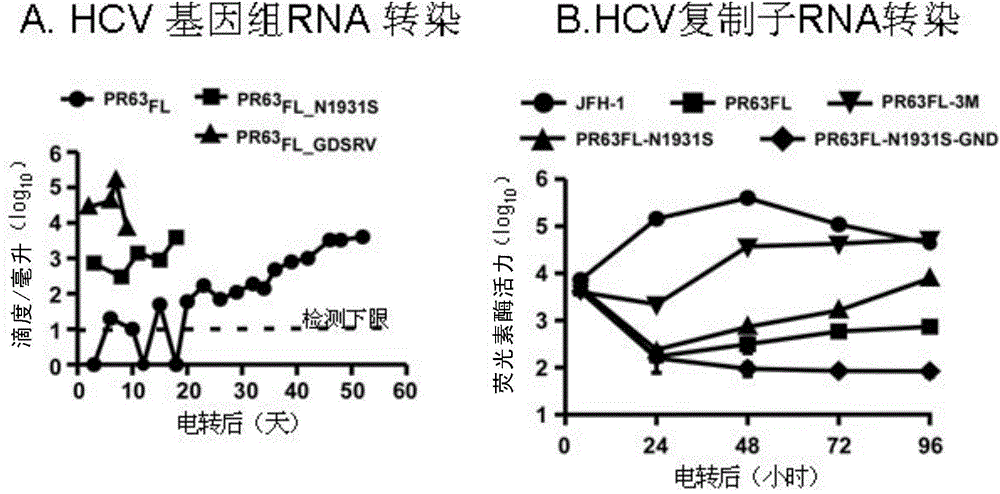
[0082] Example 4, the untranslated regions at both ends of PR63 and the C-terminus of NS5B were replaced to obtain the full-length infectious genome of PR63
[0083] Thanks to clone PR63 C-5B_A1676S_W849G_Q862P_C2432R_A2941T_S2955F / JFH-1 can effectively amplify the virus in Huh7.5.1 cells without additional mutations, based on which the inventors further narrowed down the sequence of JFH-1. First, replace the 5' untranslated region of JFH-1 origin and the N-terminal sequence of the core protein with the sequence of PR63 (replace PR63 with the 5' UTR-FspA I segment derived from PR63 C-5B_A1676S_W849G_Q862P_C2432R_A2941T_S2955F / JFH-1 genome 5'UTR-FspA I segment). The 5'untranslated region of PR63 is the same as J6CF, and there are 3 nucleotide differences compared with JFH-1. The 5'untranslated region derived from PR63 and the N-terminus of the core protein can be easily replaced without affecting the infectivity. The obtained replacement product is abbreviated as PR63 5UTR...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com