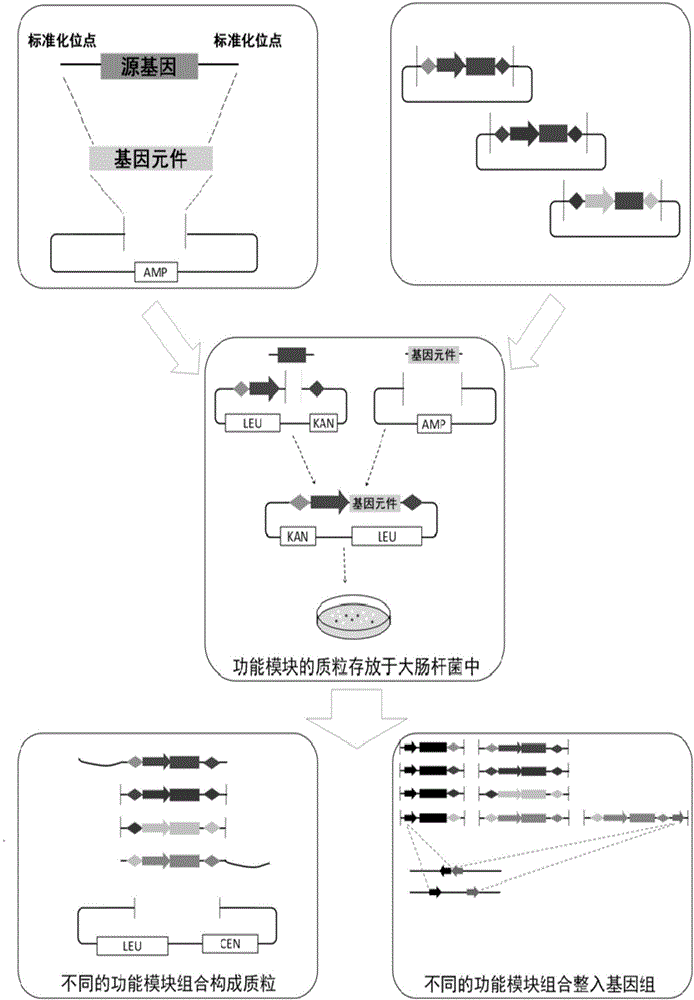
Standardized, high-accuracy and general functional module construction method
A functional module, high-precision technology, applied in the field of synthetic biology, can solve problems that do not involve the assembly of multiple modules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1 Construction of pLD-Blunt plasmid vector, pLD-Blunt plasmid vector is shown by SEQ ID NO.28.
[0044] In the present invention, pLD-Blunt is the only cloning vector artificially constructed for placing genetic elements, and its construction process is as follows:
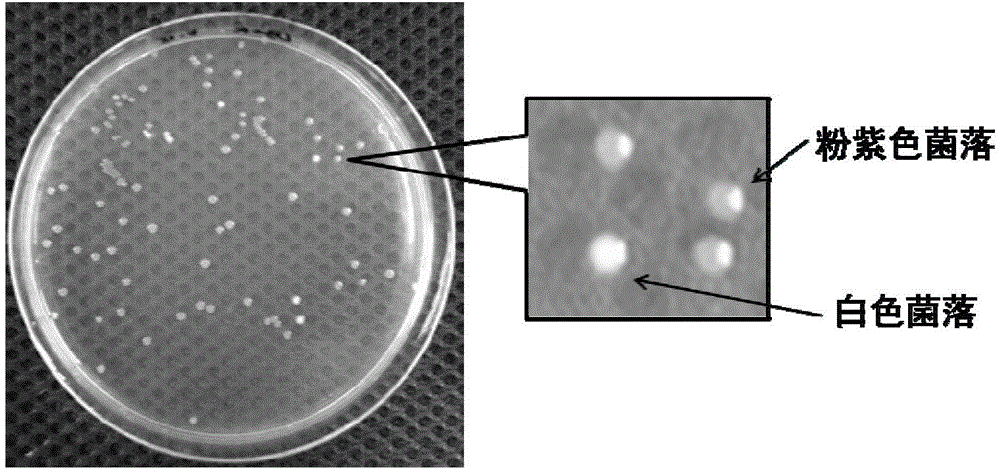
[0045](1) Select the pEASY-Blunt kit of full gold, take out 1 μL of pEASY-Blunt reagent, add 4 μL of water, react at 25°C for 5 minutes, transform Escherichia coli, pre-coat IPTG and X-gal on the plate, and use blue-white screening to obtain blue colonies.
[0046] (2) Insert the blue colony into LB-Amp liquid medium, culture overnight at 37°C, and extract the pEASY-Blunt empty plasmid vector.
[0047] (3) Use the primer pLD-Blunt1-F shown in SEQ ID NO.19 and the primer pLD-Blunt1-R shown in SEQ ID NO.20 as the upstream primer and downstream primer, and use the pEASY-Blunt empty plasmid vector as a template, The BsaI point mutation was introduced into the Amp resistance gene, and the main part of ...
Embodiment 2
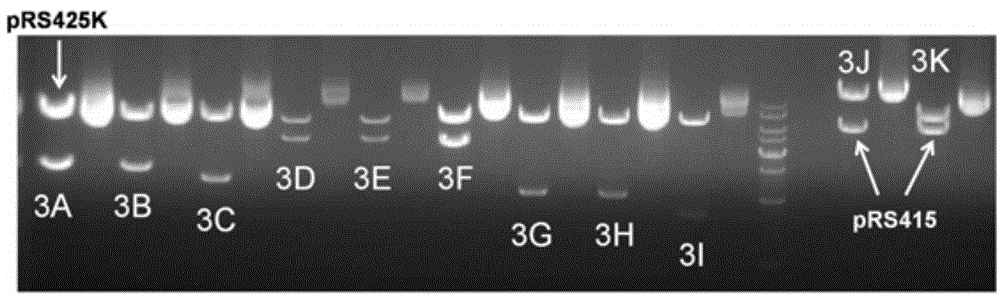
[0052] Example 2 Construction of pRS425K plasmid vector, the sequence of pRS425K is shown in SEQ ID NO.29.
[0053] In the present invention, pRS425K is an artificially constructed cloning vector for placing the module tool sequence, and its construction process is as follows:
[0054] (1) Select the pEASY-Blunt kit of full gold, take out 1 μL of pEASY-Blunt reagent, add 4 μL of water, react at 25°C for 5 minutes, transform Escherichia coli, pre-coat IPTG and X-gal on the plate, and use blue-white screening to obtain blue colonies.
[0055] (2) Insert the blue colony into LB-Amp liquid medium, culture overnight at 37°C, and extract the plasmid, which is the pEASY-Blunt empty plasmid vector. At the same time, the Escherichia coli strain containing pRS425 was inoculated to extract the pRS425 empty plasmid vector.
[0056] (3) Using the primer pRS425K1-F shown in SEQ ID NO.23 and the primer pRS425K1-R shown in SEQ ID NO.24 as the upstream primer and downstream primer, and using...
Embodiment 3
[0061] Example 3 Constructing the functional module of the green precursor deoxychromoviridans for the synthesis of violacein
[0062] Violacein is a bacteriostatic compound derived from Chromobacterium violaceum. Violacein is synthesized by cells using tryptophan as a substrate through five enzyme reactions: VioA, VioB, VioE, VioD, and VioC. After only the first three steps, the cells synthesize a green product called deoxychromoviridans, which makes yeast colonies appear green.
[0063] Therefore, in this example, three strength functional modules were designed for the genes of the three enzymes VioA, VioB, and VioE. On this basis, different functional modules were used to co-transform yeast, and different modules were combined to obtain combined functional modules. Enables Saccharomyces cerevisiae to produce deoxychromoviridans, making colonies green.
[0064] In this example, VioA is represented by SEQ ID NO.1, VioB is represented by SEQ ID NO.2, and VioE is represented ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com