Biosynthesis method of 2-deoxy scarce aldose by using aldolase
A technology of phosphate aldolase and ribose, applied in the field of biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
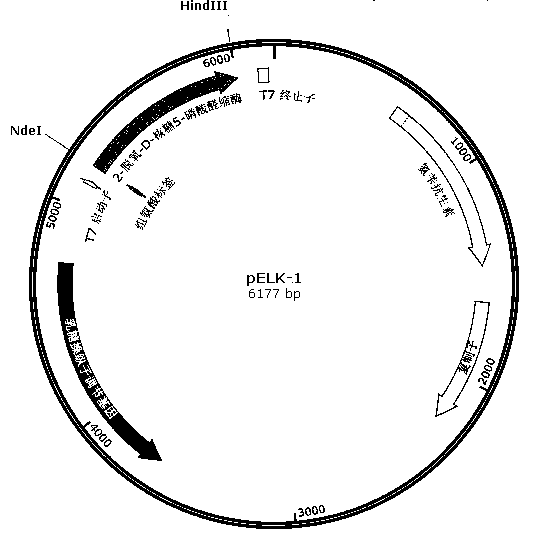
[0031] 1. Design primers 1, 2, Primer 3 and primer 4, primer 1 contains an NdeI restriction site; in primer 2 and primer 3, the 200th amino acid codon is changed from the original TTT to ATC; primer 4 has a HindIII restriction site, Moreover, the 238th amino acid codon was changed from TCC to GAC, and the 258th AGC three bases were deleted.
[0032] The primer sequences are as follows:
[0033] Primer 1: 5'-GGGTTTCATATGCATCATCACCATCACCATACTGATTTATCTGCAA
[0034] GCAGCCTG-3'
[0035] Primer 2: 5'-AAAACCGTGGGCATCAAACCGGCGGGCGGCGTGCGTACTG-3'
[0036] Primer 3: 5'-AACCGTGGGCATCAAACCGGCGGGCGGCGTG-3'
[0037] Primer 4: 5'-ACTCAAGCTTTTAGCTGCTGGCGCTCTTACCGTC-3'
[0038] 2. Using the genome of Klebsiella pneumoniae MGH 78578 as a template, use primer 1 and primer 2, primer 3 and primer 4 to amplify respectively, and obtain fragment (1) (637bp, sequence 6 in the sequence listing) and fragment (2) respectively ) (200bp, sequence 7 in the sequence listing); fragment (1) and fragment...
Embodiment 2
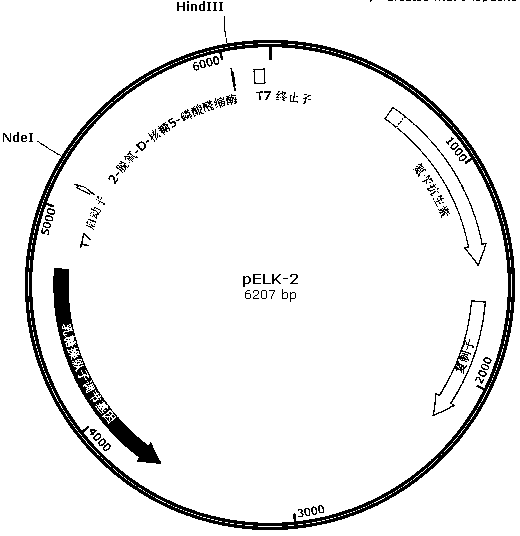
[0044] 1. According to the 2-deoxy-D-ribose 5-phosphate aldolase gene of Klebsiella pneumoniae MGH 78578 in Genbank (Genbank number: 5341458, 780bp, sequence 5 in the sequence table), design primer 5 and primer 5 band There is a HindIII enzyme cutting site, the 258th amino acid codon is changed from the original AGC to ACC; the 259th amino acid codon is changed from the original TAC to ACC; and a paragraph is added after the 259th amino acid codon Codon sequence: AAAACCCAGCTGTCCTGCACCAAATGG.
[0045] The primer sequences are as follows:
[0046] Primer 5: 5'-ACTCAAGCTTTTACCATTTGGTGCAGGACAGCTGGGTTTTGGTGGTGCTG
[0047] GCGCTCTTACCGTCGC-3'
[0048] 2. Using the Klebsiella pneumoniae MGH 78578 genome as a template, use primer 1 and primer 2, primer 3 and primer 5 to amplify respectively, and obtain fragment (1) and fragment (3) respectively (fragment (1) 637bp, sequence Sequence 6 in the list; Fragment (3) 230bp, sequence 8 in the sequence listing); Fragment (3) and Fragment (4...
Embodiment 3
[0052] 1. Cultivation and induction of Escherichia coli recombinant strains carrying pELK-1 or pELK-2 recombinant plasmids
[0053] Select LB medium (peptone (10g / L), yeast extract (5g / L), sodium chloride (10g / L), add ampicillin antibiotic (100mg / L) to the medium, at 37°C, 200rmp The Escherichia coli recombinant strain L1 was cultured under OD 600 When it reaches 0.6-0.8, add IPTG with a final concentration of 1 mmol, reduce the speed of the shaker to 120 rpm, and induce for about 20 hours.
[0054] 2. Collection and concentration of Escherichia coli recombinant strains
[0055] Centrifuge the induced Escherichia coli recombinant strain (100mL) at 4°C and 8000rmp for 15min to collect the bacteria, wash the bacteria twice with triethanolamine buffer (50mmol, pH 7.0), and finally rinse with triethanolamine buffer (50mmol, pH 7.0) Concentrate the bacterial solution to 10mL.
[0056] 3. Ultrasonic wall breaking and Ni column purification
[0057]Centrifuge the induced Escheric...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com