Spirulina platensis CPD (cyclobutane pyrimidine dimer) photolyase and application thereof
A technology of Spirulina platensis and photoreparation enzyme, which is applied in the biological field, can solve the problems of separation, purification and in vitro application difficulties, and achieves the effect of repairing DNA damage of skin cells, which is of great significance, huge social benefit and economic value.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Acquisition of optimized prokaryotic expression encoding gene of Spirulina platensis CPD photorepair enzyme
[0046] The original cDNA sequence of Spirulina platensis CPD photorepair enzyme (Gene ID: 15162397) was queried from the GeneBank database. According to the tRNA abundance data in E. coli cells, the rare triplet in the original coding gene of Spirulina platensis CPD photorepair enzyme The body codon was replaced with the corresponding codon nucleotide with the highest abundance of tRNA in E. coli cells, and finally the optimized gene coding sequence of Spirulina platensis CPD photorepair enzyme suitable for prokaryotic expression in E. coli was obtained. During the optimization process, 367 nucleotides were substituted and adjusted in the original cDNA sequence of the CPD photorepair enzyme of Spirulina platensis, accounting for 25% of the coding sequence, which has the nucleotide sequence shown in SEQ ID NO:1.
Embodiment 2
[0047] Example 2 Construction of Expression Vector of Spirulina platensis CPD Photorepair Enzyme
[0048] 1. Design primers based on the complete sequence of the optimized Spirulina platensis CPD photorepair enzyme gene:
[0049] Forward primer: CGACGCATATGGCTGCTAACCCGCAG
[0050] Reverse primer: GACACTCGAGTTAGGTCATACGCAGAGC
[0051] The PCR reaction conditions were: pre-denaturation at 98°C for 10 min, denaturation at 98°C for 50 s, annealing at 55°C for 50 s, extension at 72°C for 2 min, and the reaction was carried out for 30 cycles. The PCR reaction system is as follows:
[0052] reaction system Volume (μL) 2xPrimer star Mix 25 forward primer 1 reverse primer 1 template DNA 1 wxya 2 o 22
[0053] 2. The PCR amplification product of the optimized Spirulina platensis CPD photorepair enzyme gene and the prokaryotic expression vector pET28 were respectively digested with Nde I and Xho I at 37°C for 2 hours, and recovered with 1...
Embodiment 3
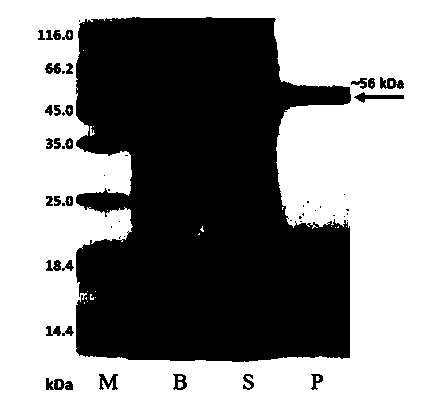
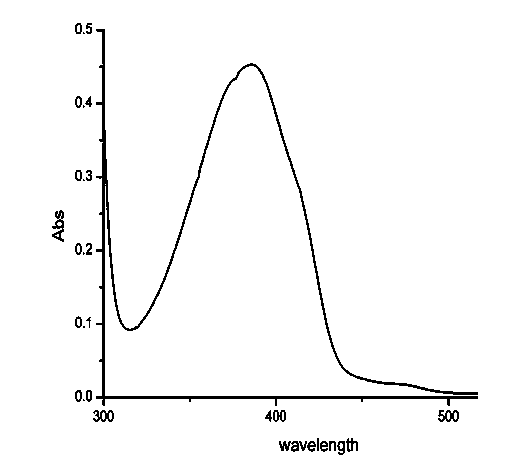
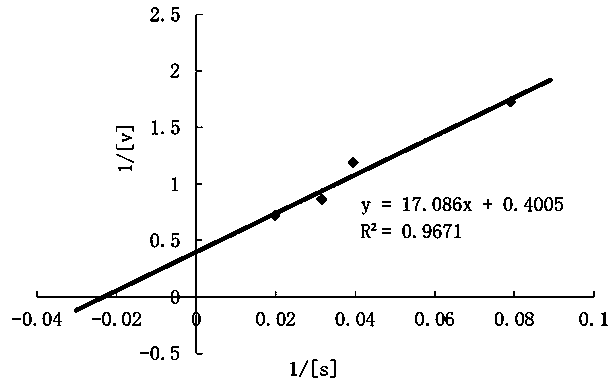
[0062] Example 3 Purification and protein detection of Spirulina platensis CPD photorepair enzyme
[0063] 1. Inoculate the recombinant strain Rosetta (DE3) / pET28 / phr in 5 mL of LB liquid medium containing kanamycin, and cultivate overnight at 37°C with shaking. The next day, take 5 mL of the bacterial liquid and insert it into 500 mL of the same medium to continue culturing for ~4h (OD 600 After about 0.8), IPTG with a final concentration of 1 mM was added, cultured with shaking at 16° C. for 20 h, centrifuged at 8000 rpm for 5 min, and the cells were collected.
[0064] 2. Ni agarose affinity column affinity chromatography: the metal chelator Ni agarose is loaded into the chromatography column, and equilibrated with 1×Ni column binding buffer (25mM Tris, 500mM NaCl, 10% Glycerol, pH 8.0). Apply the supernatant to the column. Usually eluted with 300mM imidazole to collect the target protein.
[0065] 3. Superdex 200 gel size exclusion chromatography: equilibrate the Superd...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com