Molecular Marker of Cucumber Fusarium Wilt Resistance Gene foc-4 and Its Special Primers and Application
A cucumber fusarium wilt, molecular marker technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problem of increased production input, increased technical difficulties and labor costs, and unsatisfactory chemical control effects and other problems, to achieve the effect of shortening the breeding period and improving the breeding efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1 Acquisition of Cucumber Fusarium Wilt Resistance Gene Foc-4
[0024] 1. Preliminary and fine mapping of the resistance gene Foc-4 of cucumber wilt physiological race 4
[0025] The specific positioning method includes the following steps:
[0026] A. Cucumber Fusarium wilt resistance gene mapping research Parents and genetic groups:
[0027] WI2757 (male parent) and Jinyan No. 2 (female parent) were selected as anti-susceptible parents to construct 90 RIL-F 8 strains, 130 strains of F 2:3 populations and more than 2000 strains of F 2 large groups. Among them, the male parent WI2757 and the female parent Jinyan No. 2 were purchased from the Beijing Crop Germplasm Bank.
[0028] B. Investigation of Disease Index of Cucumber Fusarium Wilt Resistance Gene Mapping Research:
[0029] The seeds of parents and each group were wrapped with gauze, soaked in warm soup, and then sowed in 50-hole hole trays after germination at a constant temperature of 28°C. Seedlin...
Embodiment 2
[0059] Example 2: Acquisition and functional verification of dCAPs marker FWSNP1 co-segregated with cucumber wilt resistance gene Foc-4
[0060] 1. Acquisition of dCAPs marker FWSNP1
[0061] It can be seen from Example 1 that there is a SNP in the Foc-4 gene between the resistant and susceptible materials, the base in the resistant material is A, and the base in the susceptible material is G, using this SNP site, according to the online design of dCAPs primers The website http: / / helix.wustl.edu / dcaps / dcaps.html, develops and designs dCAPs molecular markers closely linked to resistance / sensitivity to Fusarium wilt, named FWSNP1, and its upstream and downstream primers are shown in the sequence listing as SEQIDNO:1 and SEQIDNO: 2.
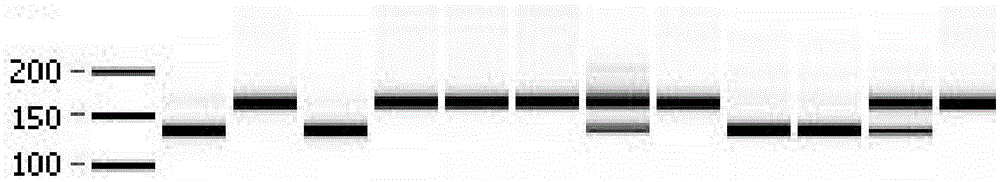
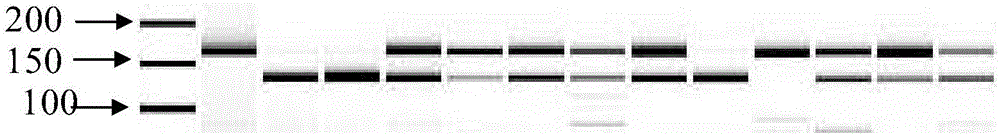
[0062] Carry out PCR amplification with the genomic DNA of parental material (resistant material WI2757 and susceptible material Jinyan No. The products were digested to obtain specific bands of resistance / susceptibility respectively, in which the...
Embodiment 3
[0068] Example 3 Identification of Fusarium Wilt Resistance of Cucumber Materials Using dCAPs Molecular Marker FWSNP1
[0069] 1. Materials to be tested: use WI2757 and Changchun Mici as parents to construct F 2 Genetic population, from which 100 individual plants were randomly selected for the following experiments. Among them, the male parent Changchun Mici and the female parent WI2757 were purchased from the Beijing Crop Germplasm Bank.
[0070] 2. Wrap the seeds of the above-mentioned materials to be tested with gauze, soak them in warm water, sow them in 50-hole trays after accelerating germination at a constant temperature of 28°C, and raise seedlings in an air-conditioned greenhouse. Extract genomic DNA from the seedlings of the material to be tested, use each genomic DNA of the material to be tested as a template, utilize upstream and downstream primers SEQIDNO:1 and SEQIDNO:2 to carry out PCR amplification, and carry out SalI endonuclease digestion identification to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com