Compositions and methods for re-programming cells without genetic modification for repairing cartilage damage
A technology of cells and substances applied in the field of compositions and methods of non-genetically modified reprogrammed cells for repairing cartilage damage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0111] Example 1 Reprogramming somatic cells into induced pluripotent stem cells (iPSCs)
[0112] 1.a. Preparation of transduction substances Oct4-11R, Sox2-11R, Klf4-11R and cMyc-llR.
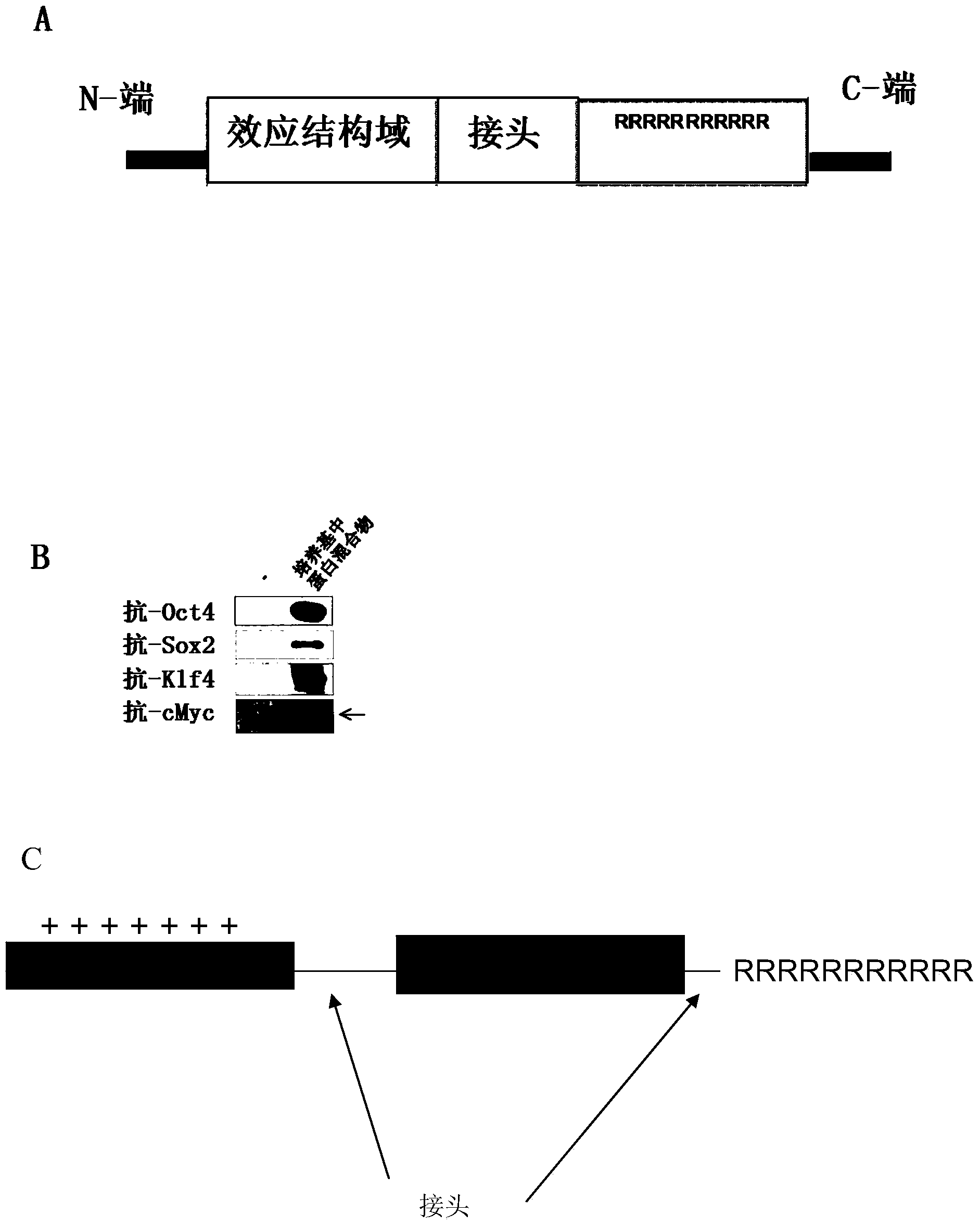
[0113] The polyarginine protein transduction domain is fused to the C-terminus of each reprogramming protein Oct4, Sox2, Klf4 and cMyc A through the linker SEQ ID NO.55 to form fusion proteins Oct4-11R, Sox2-11R, Klf4 respectively -11R and cMyc-l lR ( figure 1 A). These poly-arginine fusion proteins were expressed in E. coli in the form of inclusion bodies, which were subsequently dissolved, refolded and further purified to obtain transducible substances Oct4-11R, Sox2-11R, Klf4-11R and cMyc-11R. Protein identity was confirmed by mass spectrometry and Western blot analysis ( figure 1 B).
[0114] 1.b. Cell permeability and stability of transduction substances Oct4-11R, Sox2-11R, Klf4-11R and cMyc-11R
[0115] Add transduction substances (Oct4-11R, Sox2-11R, Klf4-11R or cMyc-11R) at differ...
Embodiment 2
[0120] Example 2 Reprogramming of liver and pancreatic exocrine cells into insulin-producing beta cells in mice by transduction substances His6-Ngn3-11R, His6-PDX1-11R and His6-MafA-11R.
[0121] The polyarginine protein transduction domain was fused to the C-terminus of each reprogramming protein (Ngn3, PDX1 and MafA) through a linker (SEQ ID NO:55) to form His6-Ngn3-11R, His6-PDX1, respectively -11R and His6-MafA-11R ( Figure 7 ). The addition of His6 (SEQ ID NO:59) facilitates protein purification. These poly-arginine fusion proteins were expressed in E. coli in the form of inclusion bodies, which were subsequently dissolved, refolded, and further purified to prepare transducible substances His6-Ngn3-11R, His6-PDX1-11R, and His6-MafA-11R. Protein identity was confirmed by mass spectrometry and Western blot analysis.
[0122] Six CD-1 mice (Charles River Laboratory) were divided into two groups: treatment group and control group. Transduce substances His6-Ngn3-11R (lmg / ...
Embodiment 3
[0123] Example 3 Reprogramming T cells with the transduction substance Foxp3 and programming them into Treg cells.
[0124] The polyarginine protein transduction domain was fused to the C-terminus of each reprogramming protein Foxp3 through a linker (SEQ ID NO:55) to form His6-Foxp3-11R ( Figure 7 ). The addition of His6 (SEQ ID NO:59) facilitates protein purification. The polyarginine fusion protein was expressed in Escherichia coli in the form of inclusion body, and then dissolved, refolded and further purified to prepare the transduction material His6-Foxp3-11R. Protein identity was confirmed by Western blot analysis.
[0125] 100 ml of healthy human blood was collected from donors, and peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation with Histopaque-1077 (Sigma-Aldrich, St Louis, MO). CD14+ monocytes were depleted by magnetic bead selection (Miltenyi Biotec, Auburn, CA). Briefly, 108 PBMCs were incubated with 200 μL of anti-C...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com