Method for extracting nucleic acid from biological sample
A technique for extracting nucleic acids from biological samples, which is applied in the field of environmental microbiology to achieve the effect of wide coverage and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] 1. Preparation before sample collection
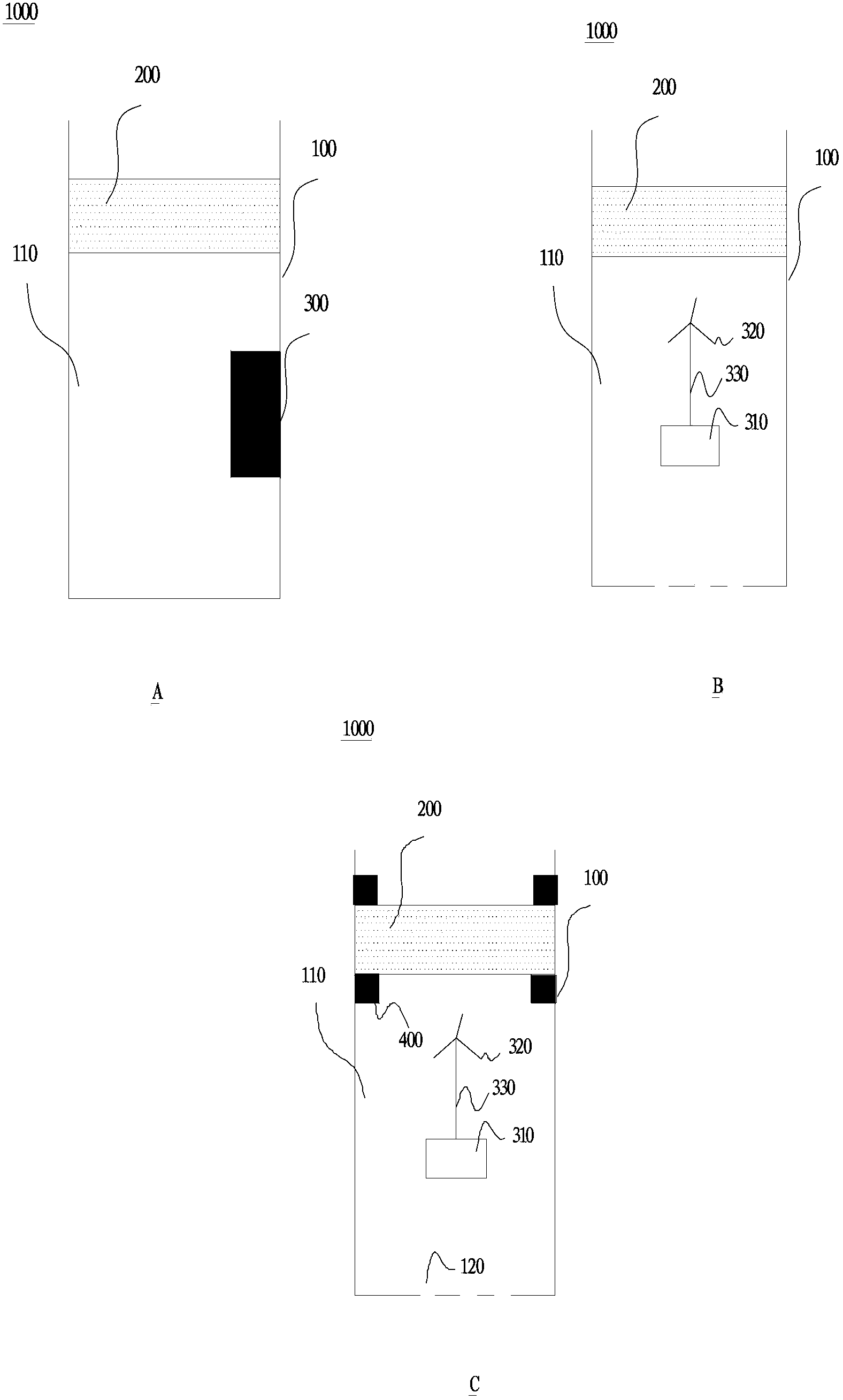
[0039] according to figure 1 C and figure 2 As shown in the structure, take a new cellulose acetate membrane, connect the filter collection assembly to the housing, and prepare the collection equipment.
[0040] 2. Sample collection
[0041] Turn on the switch of the air enrichment device, and under the pressure of 1000Pa<H≤3000Pa, start up and pump air continuously for 3 days in the workshops of the two breweries respectively. Inside, the filter membrane was torn off to collect the bacteria, and air sample 1 and air sample 2 were collected.
[0042] 3. Air sample nucleic acid extraction
[0043] Put the collected bacteria together with the torn surface filter membrane into a 50mL centrifuge tube, add 20mL TE, 200μL 10mg / mL lysozyme, 200μL 10mg / mL helicase and 200μL 10mg / mL lysozyme, blow Aspirate and mix well, incubate overnight at 37°C, digest with a mixer and mix evenly (overnight digestion); centrifuge the resulting di...
Embodiment 2
[0053] Put 0.5g of the collected bovine rumen sample into a 2mL centrifuge tube, add 500mL TE, 20μL 10mg / mL lysozyme, 20μL 10mg / mL helicase, 20μL 10mg / mL lysozyme, mix well by pipetting, and incubate overnight at 37°C , digest with a mixer (overnight digestion); centrifuge the resulting digestion solution at 4°C at 9000r / min for 10 minutes, remove the supernatant, and keep the precipitate; add 400 μ TENS lysate (200 mmol / L NaCl, 100mmol / L Tris-HCl, pH 8.0, 2.0%SDS, 50mmol / L EDTA, 0.5%Triton X-100) and 20mg / mL proteinase K 300μL, invert and mix well, incubate at 55℃ for 1h, invert and mix every 15min Homogenize once; Centrifuge the lysed sample at 10000r / min at 4°C for 10min, keep the supernatant, discard the precipitate; add an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1) to the supernatant , mix well, and centrifuge at 12000r / min at 4°C for 10min; transfer the obtained supernatant to a new tube, add an equal volume of chloroform:isoamyl alcohol (24:1) to it, mi...
Embodiment 3
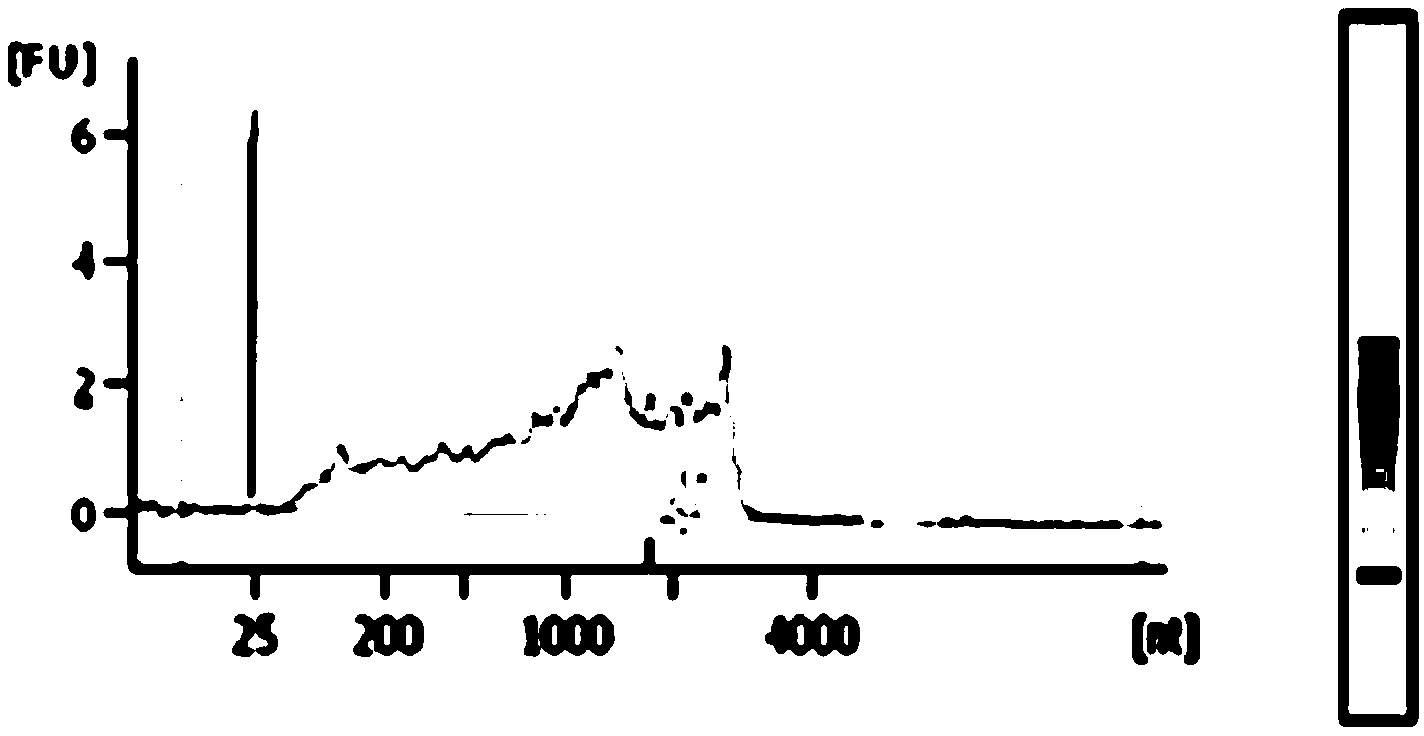
[0055] Adopt dental plaque sample, repeat embodiment 2 to extract DNA, gained experimental result sees Figure 8 .
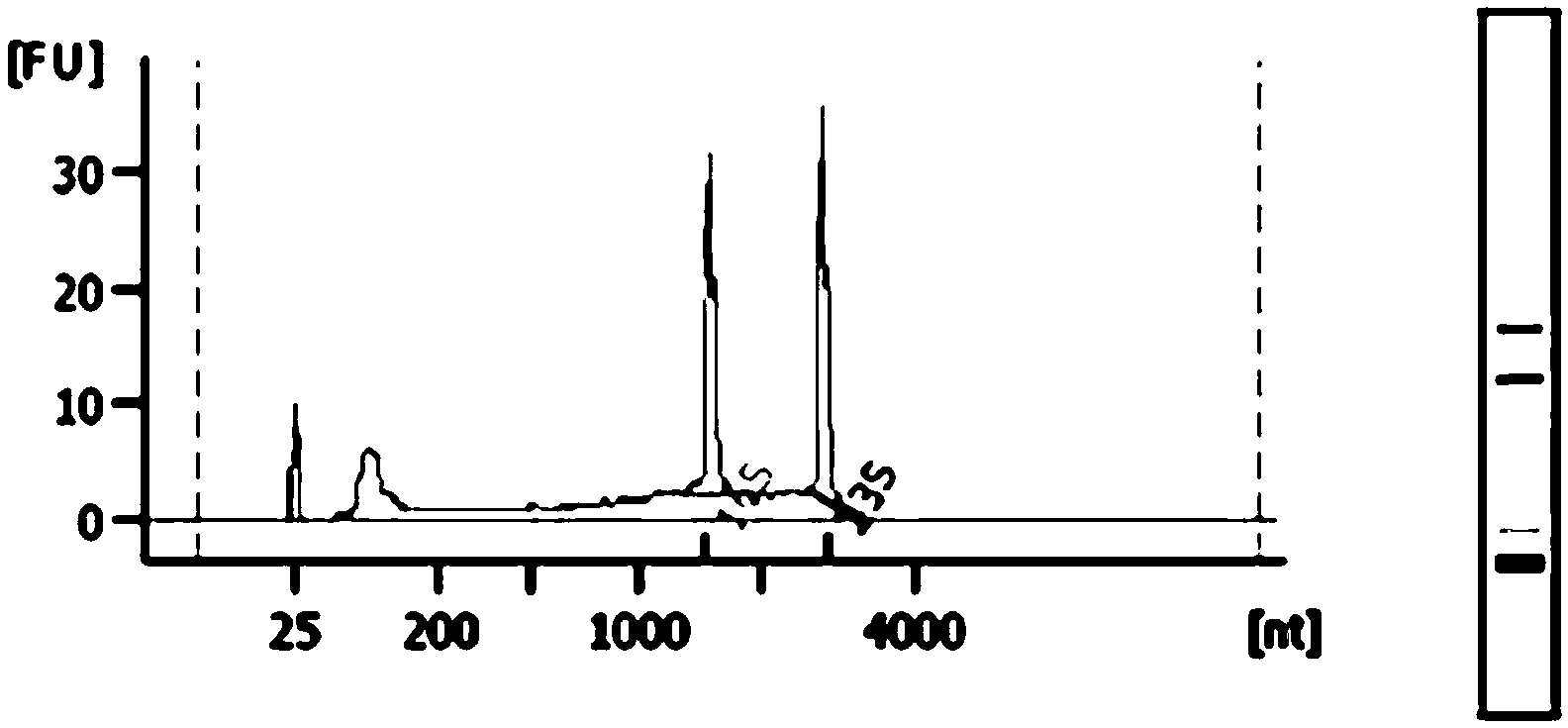
[0056] like Figure 7 and Figure 8 As shown, using the method of the present invention, nucleic acid samples can be effectively extracted from microorganisms.
PUM
| Property | Measurement | Unit |
|---|---|---|
| pore size | aaaaa | aaaaa |
| pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com