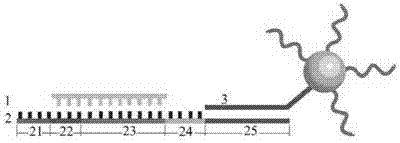
DNA molecular machine and single-base mutation detection method based on DNA molecular machine as well as application of method
A DNA molecule and single base mutation technology, applied in the field of DNA molecular detection, to achieve the effect of simple detection means and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] In the first step, probe chain 1 and probe chain 2 are modified on the surface of gold nanoparticles by using the reaction between thiol and gold:
[0037] ①Mix the oligonucleotide and 13nm nano-gold with a molar concentration ratio of 350:1, and store at 4°C for 16 hours;
[0038] ② Add final concentration of 10mM phosphate buffer and store at 4°C for 12 hours;
[0039] ③Add concentrated NaCl solution to change the final concentration from 0.1M to 0.2M and then to 0.3M, and store at 4°C for 8 hours at each concentration;
[0040] ④Centrifuge at 13000rpm for 30 minutes, discard the supernatant;
[0041] ⑤Add 10mM phosphate buffer, wash twice with final concentration of 0.1M NaCl;
[0042] ⑥Add 10mM PBS, 0.1M NaCl, pH=7.4, store at 4°C.
[0043] ⑦The concentration of 13nm nano-gold solution is determined by the ultraviolet absorption peak at 520nm wavelength.
[0044] The second step, annealing to prepare binary hybrid strands:
[0045] a. Dissolve the connecting chai...
Embodiment 2
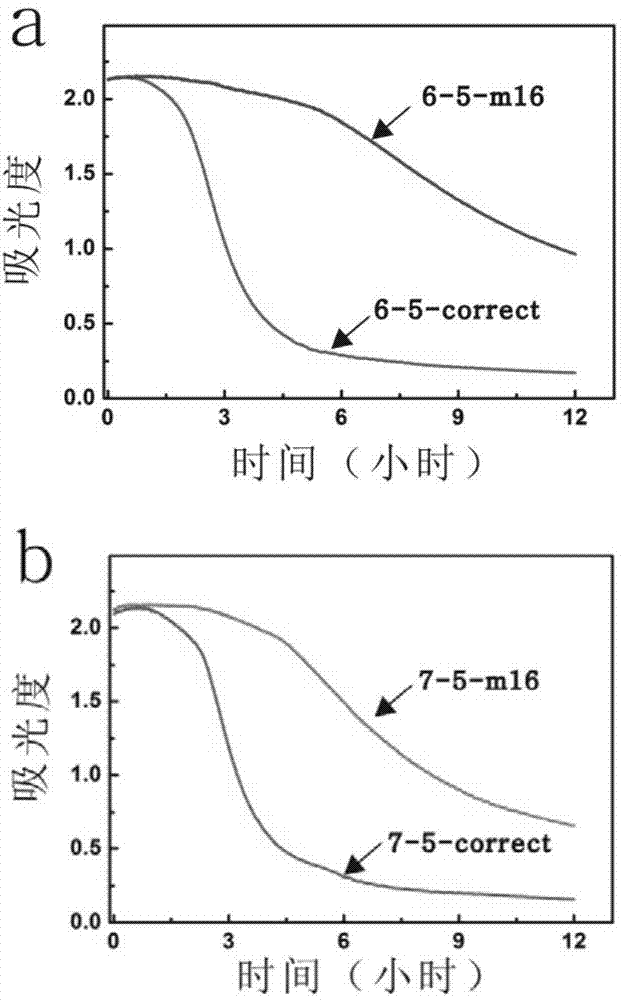
[0068] Adopt the same operating steps as in Example 1, but reduce the 5' end of the protective chain by one base, resulting in a change in the structure of the DNA molecular machine, 22=5, 24=6 in the example 1, in this example 2 District=5, District 24=7. It is also possible to effectively distinguish the presence of base mutations in about 4 hours. UV dynamic process see figure 2 b.
[0069] Sequence of connecting strands (5'-3' from left to right)
[0070] CATCCCACTCCACCTTCATCTCACTACGATCCTCACTCTCACCCTAC
[0071] Sequence of protective strands (5'—3' from left to right)
[0072] AGTGAGATGAAGGTGGAGTG
[0073] Sequence of thiol-modified probe 1 (5'—3' from left to right)
[0074] SH-TTTTTTTTTTTTTTGTAGGGTGAGAGTGA
[0075] Sequence of thiol-modified probe 2 (5'—3' from left to right)
[0076] SH-TTTTTTTTTTTTTTGTAGTGAGATGAAGGTGGAGTGGGATG
[0077] Fully complementary catalytic strand sequence (5'-3' from left to right)
[0078] AGATCGTAGTGAGATGAAGGTG
[0079] Sequences...
Embodiment 3
[0082] The same operation steps as in Example 1 were adopted, except that the gold nanoparticles modified by the probe chain 1 were mixed with the binary hybridization chain at a molar ratio of 1:21. The presence of base mutations can be effectively distinguished when the reaction is carried out for about 4 hours. UV dynamic process see figure 2 c.
[0083] Sequence of connecting strands (5'-3' from left to right)
[0084] CATCCCACTCCACCTTCATCTCACTACGATCCTCACTCTCACCCTAC
[0085] Sequence of protective strands (5'—3' from left to right)
[0086] TAGTGAGATGAAGGTGGAGTG
[0087] Sequence of thiol-modified probe 1 (5'—3' from left to right)
[0088] SH-TTTTTTTTTTTTTTGTAGGGTGAGAGTGA
[0089] Sequence of thiol-modified probe 2 (5'—3' from left to right)
[0090] SH-TTTTTTTTTTTTTTGTAGTGAGATGAAGGTGGAGTGGGATG
[0091] Fully complementary catalytic strand sequence (5'-3' from left to right)
[0092] AGATCGTAGTGAGATGAAGGTG
[0093] Sequences with single base mutations (5'-3' fr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com