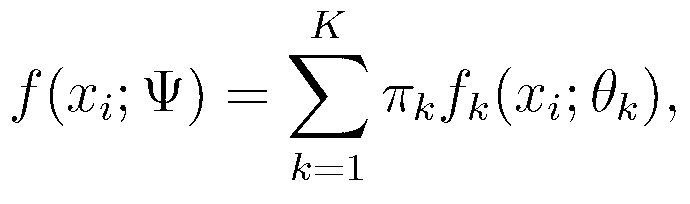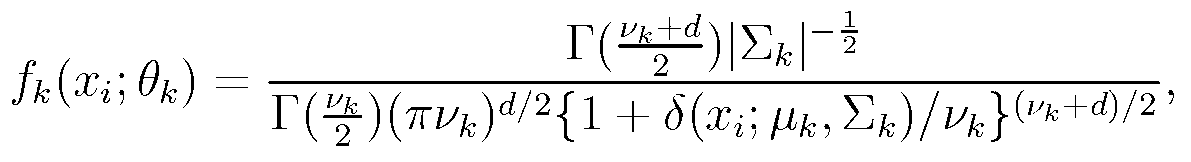Cancer hypotype biomarker detecting system based on student t distribution
A technology for biomarkers and detection systems, applied in the field of cancer subtype biomarker detection systems, can solve the problems of ignoring inter-gene correlations and unfavorable understanding of collective biological functions of genes.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
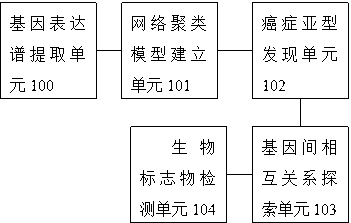
[0083] Such as figure 1 As shown, it is a structure diagram of an embodiment of a cancer subtype biomarker detection system based on Student's t distribution of the present invention. see figure 1 , a cancer subtype biomarker detection system based on Student's t distribution in this embodiment, comprising:
[0084] The gene expression profile extraction unit 100 extracts the expression profiles of n independent samples containing d genes from the gene chip X={x 1 ,...,x n}, where x i =[x i1 x i2 … x id ] represents the expression profile of d genes in sample i; each gene is pre-standardized, the sample mean is 0, and the sample variance is 1.
[0085] The network clustering model building unit 101 is connected with the gene expression profile extraction unit 100, and the extracted gene expression profile X={x 1 ,...,x n}Input it, establish a mixture model and solve the mixture model, wherein the mixture model uses a multivariate Student's t distribution to describe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com