Method for remaking multiple cloning sites of known vector
A technology of multiple cloning sites and vectors, applied in the field of cloning, can solve problems such as low operational flexibility, low construction efficiency, and low yield of fragments, and achieve high flexibility, easy operation, and high efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
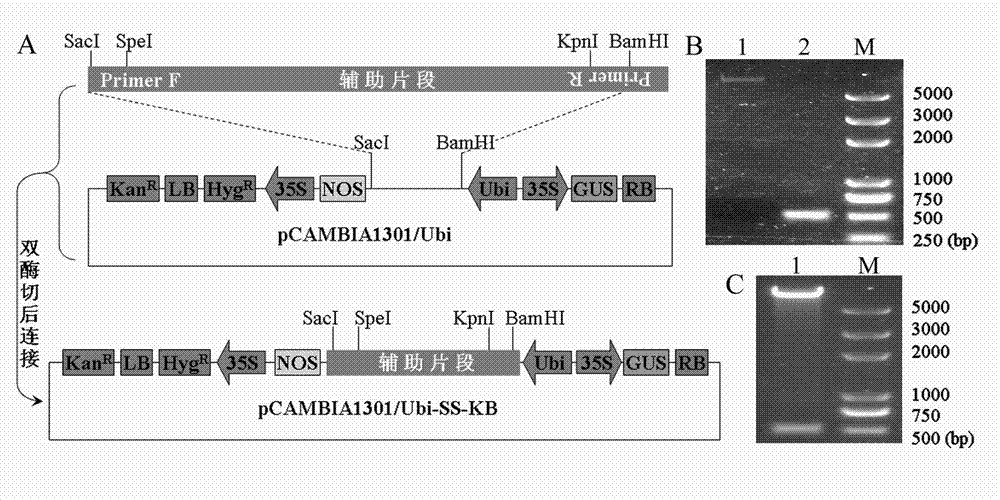
[0023] Embodiment 1: Transformation of multiple cloning sites of plant expression vector pCAMBIA1301 / Ubi
[0024] In the research work of this experiment, a fragment of 8.2kb needs to be connected into the overexpression vector pCAMBIA1301 / Ubi, and the analysis of the restriction site shows that the restriction site suitable for this fragment is only Kpn I and Speech 1, and the available enzyme cutting point on the overexpression vector pCAMBIA1301 / Ubi is Bam H I and Sac I, in order to successfully connect the target fragment into the vector, the adapter primer-PCR method will Kpn I and Speech The I restriction site was introduced into the multiple cloning site of the pCAMBIA1301 / Ubi vector.
[0025] 1) Design of adapter primer pairs
[0026] Using a pair of existing primer pair SS with better amplification efficiency, namely SS-F: 5'ACCTATCACCAAATGTACCC3', SS-R: 5'CAATCCTCAATATGCTCAAC3' as the basic primers, the primer pair SS is derived from rice genomic DNA,...
specific Embodiment approach 2
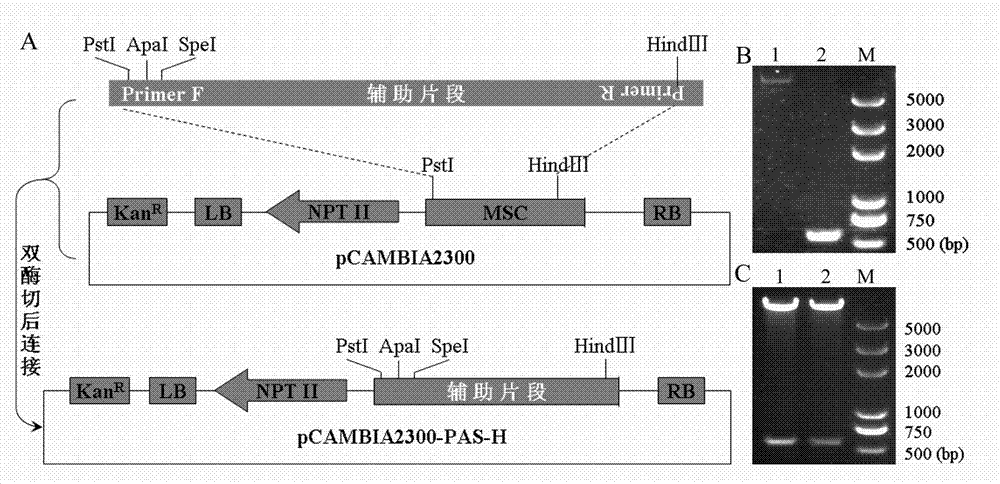
[0029] Specific embodiment two: Transformation of multiple cloning sites of plant expression vector pCAMBIA2300
[0030] In another research work, we need to use the Hind III and Apa I and contain Hind III and Speech I The pCAMBIA2300 vector of the restriction site, we also transformed the known multiple cloning site of pCAMBIA2300 by the adapter primer-PCR method.
[0031] 1) Design of adapter primers
[0032] A pair of existing primer pair H with better amplification efficiency was used as basic primers, namely H-F: 5'TCATGGAGTCAAAGATTC3', H-R: 5'AGTCCCCCGTGTTCTCTC3', and a pair of adapter primer pair H-SAP was designed. The basic primers were derived from Escherichia coli 35S promoter sequence, the size of the amplified fragment is 569bp, and the amplified sequence does not contain Hind III and Pst I digestion recognition sequence. We add Hind III Restriction site sequence and protective bases, the primer sequence before the adapter is obtained, which is...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com