Non-exogenous induced factor integrated porcine induced pluripotent stem cell and its construction method
A technology of pluripotent stem cells and construction methods, applied in the field of induced pluripotent stem cells, porcine induced pluripotent stem cells and their construction, can solve the problems of increasing the risk of carcinogenesis of iPS cells, and achieve the effect of reducing the risk of carcinogenesis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1, construction of lentiviral vector Lv-ef1α-Cre
[0039]1.1. Design Cre primers according to the sequence of pBluescript II SK(-)-Cre (Stratagene), and introduce restriction sites. The primer sequences are shown in Table 1 (wherein F represents the forward primer and R represents the reverse primer).
[0040] Table 1 Primer Sequence
[0041]
[0042] Note: The lowercase letters in the primer sequences are the restriction restriction sites introduced.
[0043] 1.2PCR amplification
[0044] Using the pBluescript II SK(-)-Cre plasmid as a template, use the primers in Table 1 for PCR amplification, as follows:
[0045] Reaction system (25 μl): template 0.3 μl, forward and reverse primers 0.25 μl, 10×pfx Mix 2.5 μl, Accuprime pfx enzyme 0.2 μl, double distilled water 21.5 μl.
[0046] Reaction conditions: 95°C for 2min; 95°C for 15sec, 66°C for 20sec, 68°C for 1min, 35 cycles; 68°C for 10min.
[0047] 1.3. PCR product purification, enzyme digestion, ligation ...
Embodiment 2
[0051] Example 2, construction of lentiviral vector Lv-ef1α-CreER
[0052] 2.1. Design CreER primers according to the sequence of pCAGG-CreER (Stratagene), and introduce restriction sites. The primer sequences are shown in Table 1 (wherein F represents the forward primer and R represents the reverse primer).
[0053] 2.2. PCR amplification
[0054] Using the pCAGG-CreER plasmid as a template, PCR amplification was performed using the primers in Table 1, as follows:
[0055] Reaction system (25ul): template 0.3μl, forward and reverse primers 0.25μl each, 10×pfx Mix 2.5μl, Accuprime pfx enzyme 0.2μl, double distilled water 21.5μl.
[0056] Reaction conditions: 95°C for 2min; 95°C for 15sec, 66°C for 20sec, 68°C for 2min, 35 cycles; 68°C for 10min.
[0057] 2.3. PCR product purification, enzyme digestion, ligation
[0058] The resulting PCR product was subjected to agarose gel electrophoresis to determine its concentration. Use the universal DNA purification and recovery kit ...
Embodiment 3
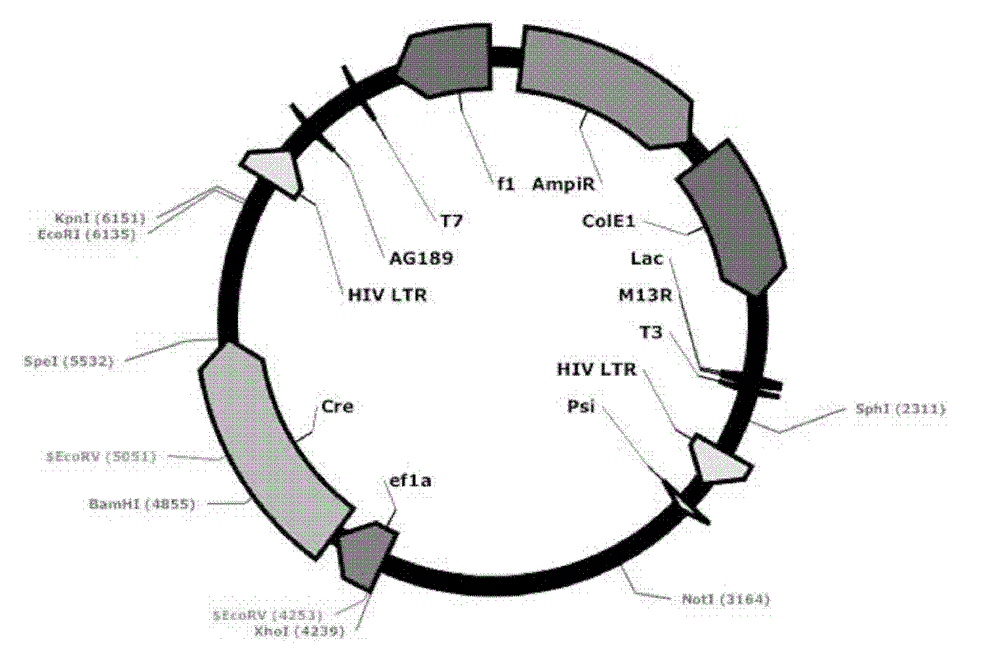
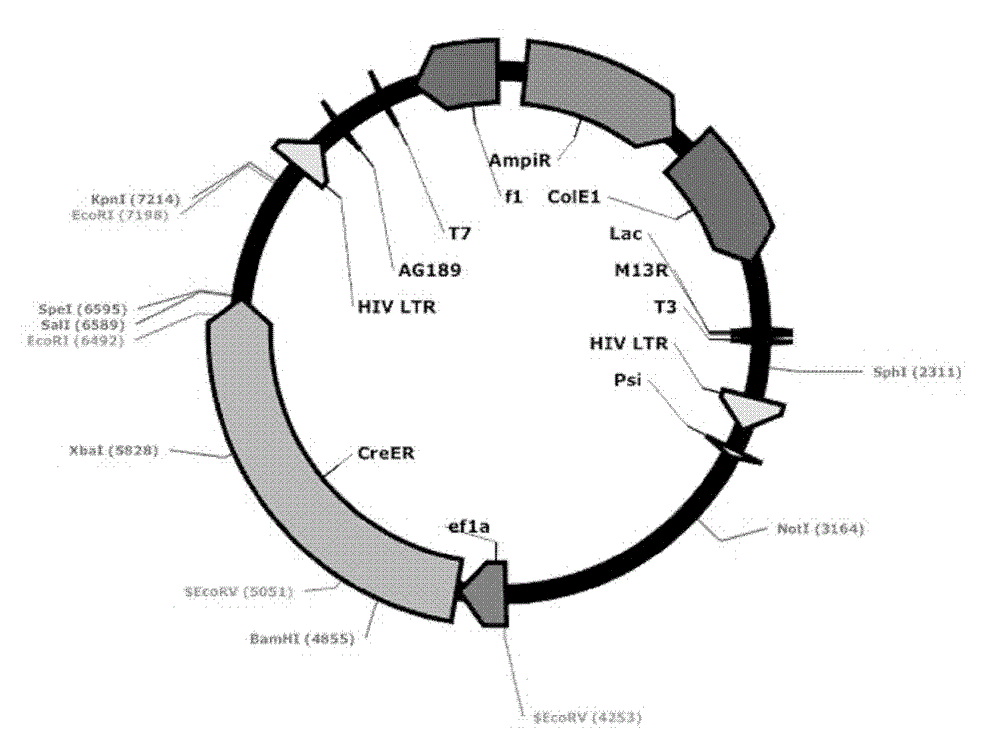
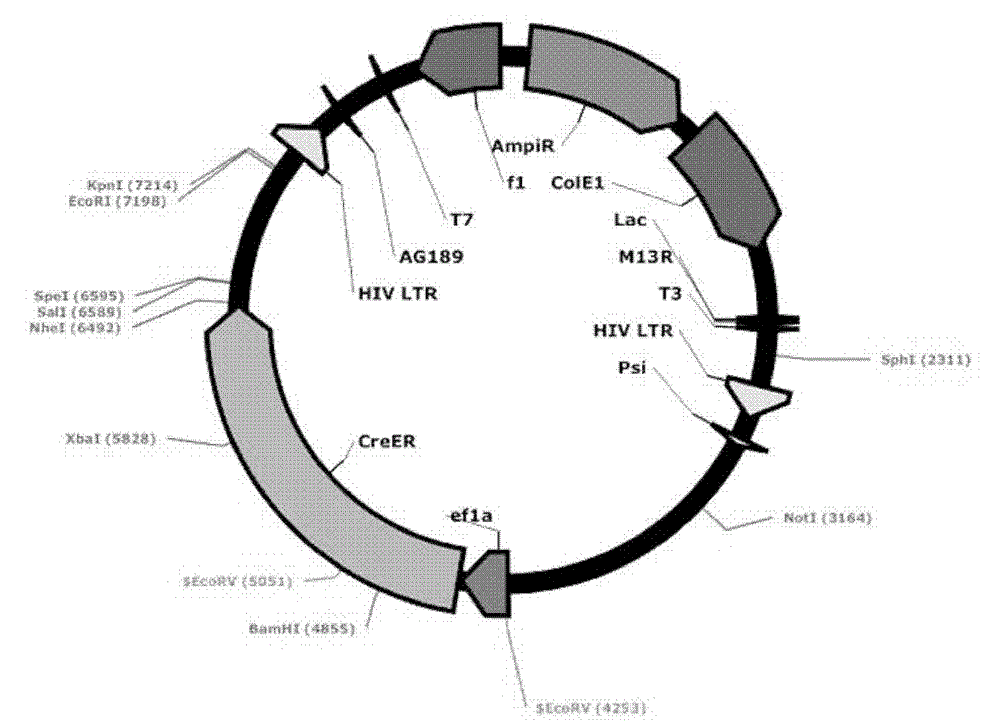
[0060] Transform the ligation product into GBE180 competent bacteria, spread it evenly on the agar plate containing ampicillin antibiotics, and cultivate it at 37°C for 16 hours, pick an appropriate number (10-20) of single colonies from the agar plate, and put it on the agar plate containing Shake culture in a 15ml round bottom centrifuge tube with 5ml LB medium for 16 hours. Use the Biotech B-type Plasmid Small Quantity Rapid Extraction Kit to extract the plasmid, use BamH I and Sal I double enzyme digestion to identify, select the plasmid with the correct digestion and send it to the sequencing company for sequencing, after sequencing identification, the correct Lv-ef1α-CreER was obtained , with a structure such as figure 2 As shown, 8539bp. Example 3. Mutation of the EcoR I restriction site behind the ER in the lentiviral vector Lv-ef1α-CreER to Nhe I
[0061] 3.1. PCR mutation primer design
[0062] In the Lv-ef1α-CreER sequence, about 20 bp were designed on both side...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com