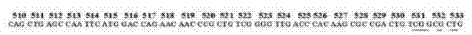Visualized detection probes and application thereof in tuberculosis rifampicin resistance mutation detection
A technology for detecting probes and mutation detection, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., and can solve problems such as complex operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] 1. Design of DNA peroxidase mutation detection probe system
[0048] (1) The sequence of the core region of the rpoB gene mutation is as follows: figure 1 shown
[0049] The present invention comprises 8 kinds of mutation types of 530 to 533 amino acids ( figure 1 underlined), the specific mutations and corresponding detection probes are as follows:
[0050] mutation site
Corresponding detection probe
531
T→G
I-B531-1C
531
C→G
I-B531-2C
[0051] 531
C→T
I-B531-2A
532
C→A
I-B532-2T
532
C→T
I-B532-2A
533
C→T
I-B533-1A
533
T→C
I-B533-2G
531
C → A, G → C
I-B531-2T3G
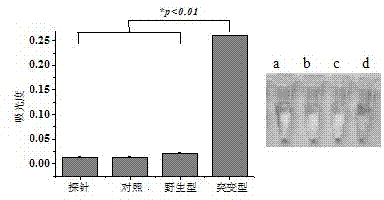
[0052] (2) Design the corresponding detection probe cluster according to the mutation site in rifampicin (RFP) resistance-determining region of Mycobacterium tuberculosis rpoB gene. The probe cluster consists of three parts: a ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com