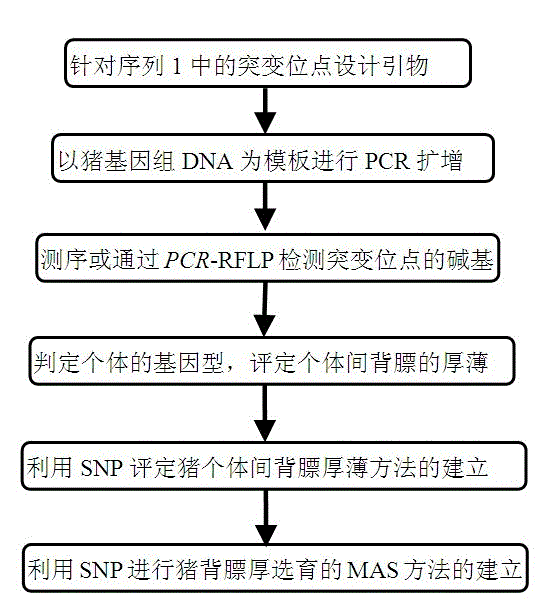
Molecule labeling method for maker-assisted selection of pig backfat thickness
A marker-assisted selection and molecular marker technology, applied in biochemical equipment and methods, microbial determination/inspection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1 Acquisition and polymorphism analysis of the DNA sequence where the SNP is used to assess the thickness of pig backfat
[0033] The DNA sample used for the SNP detection comes from the genome of the pig, which can be collected by collecting the saliva or hair follicles of the pig and then referring to the conventional method to extract the genomic DNA and then store it at -20°C for future use.
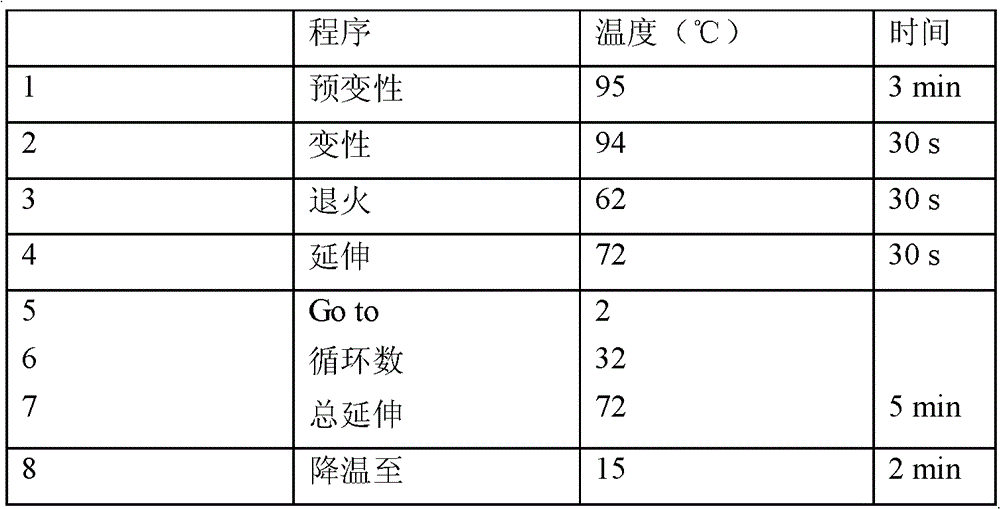
[0034] Primers PCAL / PCAR were designed according to the sequence 1 in the sequence listing, they are respectively located in the second display (Exon 2) and the third exon (Exon 3) regions of the gene, and the primer sequences are as follows:
[0035] SEQ ID NO: 3:
[0036] PCAL: 5'-GATTGAAGCGCTGCAGGAAGTC-3'
[0037] SEQ ID NO: 4:
[0038] PCAR: 5'-CAGGAGGAAGGAATTGCAGGAG-3'
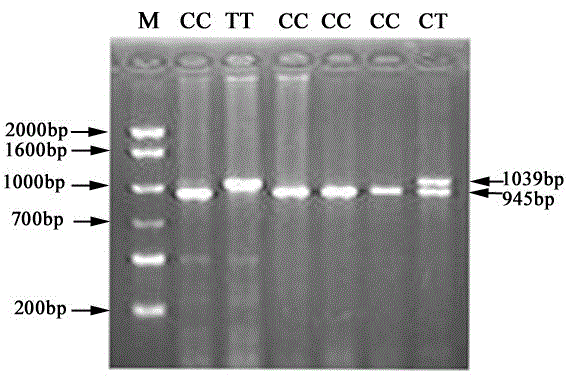
[0039] The length of the primer amplified fragment is 1039bp, which is located on the second intron of the pig CART gene, that is, the nucleotide fragment at the 5' end of the sequence in SEQ I...
Embodiment 2
[0052] Example 2 Landrace × Blue Tang F 2 Association analysis of marker genotypes and traits in resource populations
[0053] The DNA sample used for the detection of this single nucleotide polymorphism came from Changbai×Lantang F 2 Resource population, a total of 230 DNA samples. Genomic DNA was extracted by conventional methods and stored at -20°C for future use.
[0054] First, establish the following analysis model to eliminate the influence of gender, weight and batch on phenotype values:
[0055] the y ijkl =μ+s i +b j +g k + r i Cov W +e ijkl
[0056] Among them, y ijkl is the observed value of the trait, μ is the least square mean, s i is the sex effect (i=1 is boar, i=2 is sow), b j is the batch effect (j=1~4), g k Genotype effect (k = CC, CT and TT), r i is the regression coefficient of the covariate, Cov w is the covariate of live body weight before slaughter, e ijkl for the error. Assumed to obey the N (0, σ2) distribution.
[0057] Applying th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com