Molecular marker for assisted selective breeding of upland cotton with greensickness resistant traits
A Verticillium wilt resistance and assisted selection technology, applied in the field of cotton genetics and breeding, can solve the problems of no field breeding groups, etc., and achieve the effects of improving efficiency and selection accuracy, improving efficiency and accuracy, and reducing workload
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
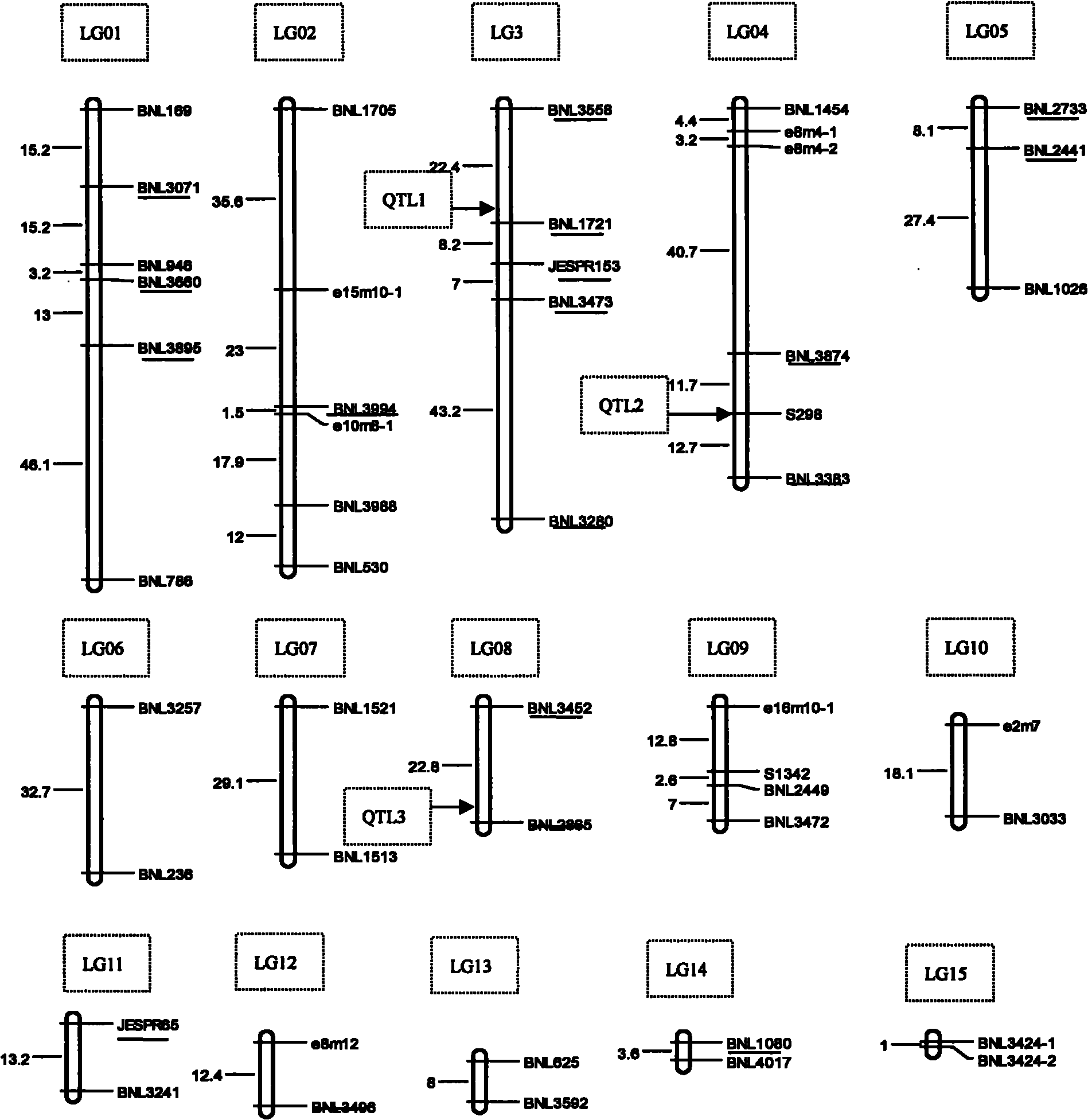
[0025] 1. Get 96 upland cotton lines (such as figure 2 Shown), with Changkang cotton and TM-1 as contrast respectively, described material is planted in Verticillium wilt disease nursery, single-row area, repeats twice;
[0026] 2. Inoculate Verticillium dahliae of the Anyang strain with moderate pathogenicity before sowing, and when the incidence of Verticillium dahliae peaks in June, July, and August, the rate of susceptible control strains reaches 80% or the disease index reaches about 50 , investigate the incidence of each test material, adopt the 5-level system to grade cotton Verticillium wilt (Table 1), and calculate the disease index and relative disease index, and finally calculate the average according to the material as the average disease index. The formulas for calculating the condition index and the relative condition index are as follows:
[0027] Disease index DI = [(number of first-level diseased plants × 1 + number of second-level diseased plants × 2 + numb...
Embodiment 2
[0048] 1. Take different upland cotton lines and plant them in the field (such as figure 2 ).
[0049] 2. Extracting its total DNA from the tender green leaves on the field material plants.
[0050] 3. Use the disease nursery to identify the Verticillium wilt resistance of the upland cotton strains, see Example 1 for the specific identification method.
[0051] 4. Utilize single SSR primer sequence of the present invention to analyze, concrete steps are as follows:
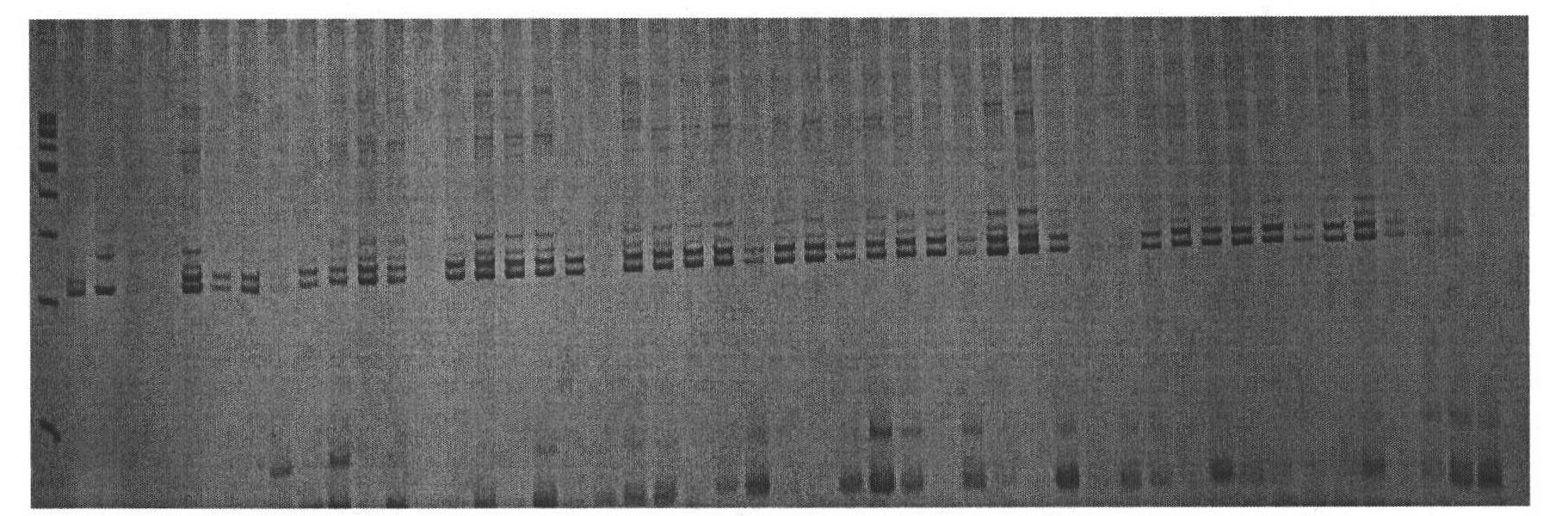
[0052] 4.1 SSR primer labeling analysis:
[0053] PCR amplification reaction system is 10 μL: 25ngDNA, 1μM upstream primer, 1μM downstream primer, 1×buffer (67mmol / L Tris-HCl, pH8.8, 16mmol / L (NH 4 ) 2 SO 4 ), 0.2mmol / L dNTPs, 2.5mmol / L MgCl 2 ,, TaqDNA polymerase 0.5U.
[0054] PCR reaction program: denaturation at 95°C for 3min; denaturation at 94°C for 45s, annealing at 60°C for 45s, extension at 72°C for 1min, a total of 30 cycles; extension at 72°C for 3min. Store at 10°C. PCR amplification was carr...
Embodiment 3
[0068] 1. Take different upland cotton strains (such as figure 2 ) planted in the field.
[0069] 2. Extracting its total DNA from the tender green leaves on the field material plants.
[0070] 3. Use the disease nursery to identify the Verticillium wilt resistance of the upland cotton strains, see Example 1 for the specific identification method.
[0071] 4. comprehensively utilize three of the present invention SSR primer sequence For analysis, the specific steps are as follows:
[0072] 4.1 SSR primer labeling analysis:
[0073] PCR amplification reaction system is 10 μL: 25ngDNA, 1μM upstream primer, 1μM downstream primer, 1×buffer (67mmol / L Tris-HCl, pH8.8, 16mmol / L (NH 4 ) 2 SO 4 ), 0.2mmol / L dNTPs, 2.5mmol / L MgCl 2 ,, TaqDNA polymerase 0.5U.
[0074] PCR reaction program: denaturation at 95°C for 3min; denaturation at 94°C for 45s, annealing at 60°C for 45s, extension at 72°C for 1min, a total of 30 cycles; extension at 72°C for 3min. Store at 10°C. PCR ampl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com