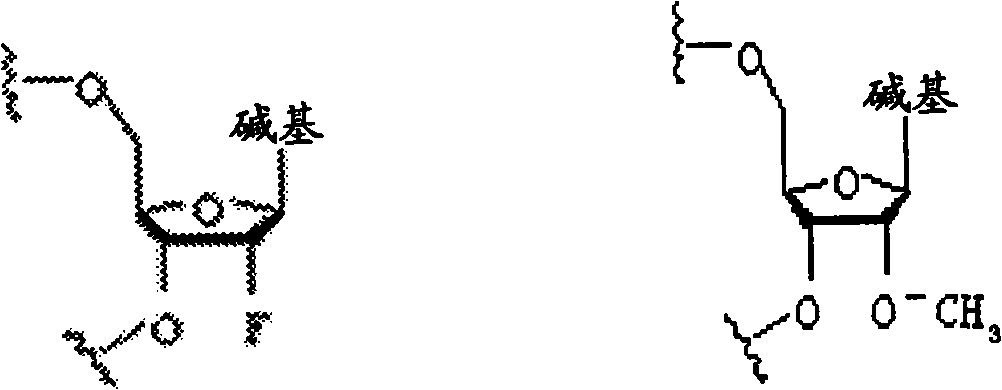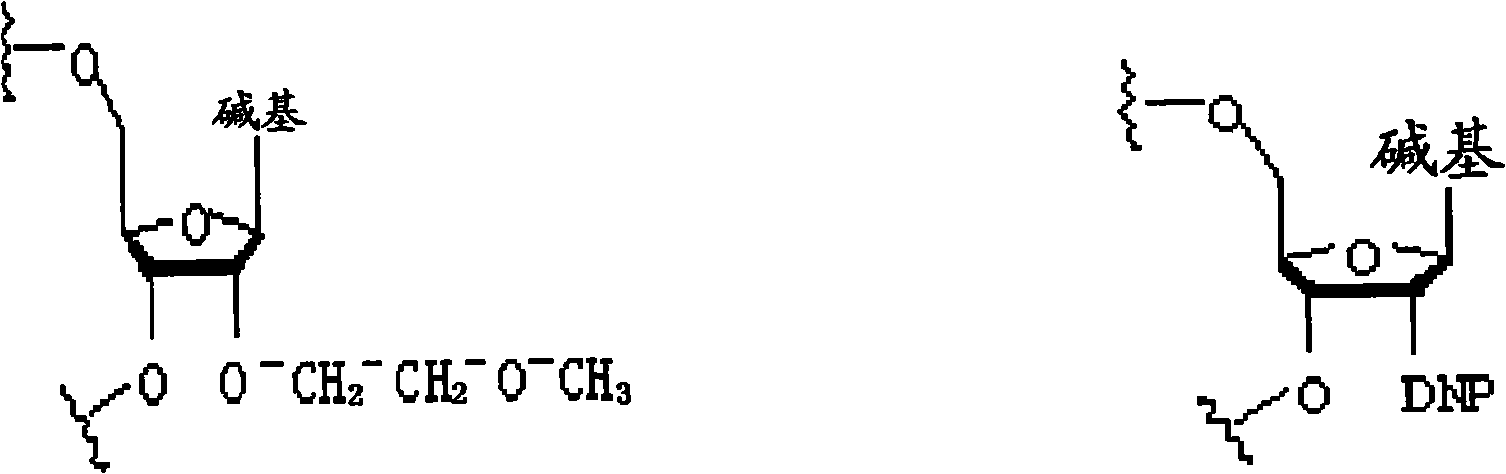Specific modification for inhibiting RNA interference off-target effect
A small interference and chemical modification technology, applied in the field of nucleic acids, can solve the problems of misinterpreting the inhibitory activity of siRNA, the toxic and side effects of siRNA drugs, and affecting the application of RNA interference technology, so as to reduce off-target effects and improve specificity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0064] Generally speaking, the synthetic method of oligonucleotide molecule comprises the following four steps: (1) synthesis of oligoribonucleotide; (2) deprotection; (3) purification and separation; (4) desalting.
[0065] For example, the chemical synthesis steps of siRNA having the nucleotide sequence shown in SEQ ID NO: 1 are as follows:
[0066] (1) Synthesis of siRNA constituent strands: set the sense strand or antisense strand oligonucleotide sequence of synthetic 1 millimolar amount of siRNA on the automatic DNA / RNA synthesizer (for example, Applied Biosystems EXPEDITE8909), set each The coupling time of each cycle is 10-15 minutes. The starting material is 5'-O-dimethoxy-thymidine support connected to the solid phase. The first cycle connects a base to the solid phase support. , and then in the nth (2≤n≤35) cycle, a base is connected to the base connected in the n-1th cycle, and this cycle is repeated until the synthesis of the entire nucleic acid sequence is complet...
Embodiment 1
[0097] Example 1. siRNA target sites and siRNA sequences
[0098] Invitrogen Beijing Branch was commissioned to synthesize the siRNA target sequence fragments in Table 1 for the construction of fusion reporter gene expression vectors.
[0099] Design siRNA for the target site sequence in Table 1, and entrust Guangzhou Ruibo Biotechnology Co., Ltd. to synthesize the chemically modified and unmodified small interfering RNA (siRNA) listed in Table 2-11.
[0100] Table 1. siRNA target site sequences
[0101]
[0102]
[0103]
[0104]
[0105]
[0106]
[0107]
[0108]
[0109]
[0110]
[0111]
[0112]
[0113]
[0114]
[0115]
[0116]
[0117]
[0118]
[0119]
[0120]
[0121]
[0122]
[0123]
[0124]
Embodiment 2
[0125] Example 2. Detection of siRNA inhibitory activity on target sites
[0126] 1) Construction of recombinant firefly luciferase reporter gene plasmid with siRNA isolation target site
[0127] Refer to the following literature to construct a recombinant firefly luciferase reporter gene plasmid containing siRNA isolation target sites. The reporter gene plasmid vector can be obtained from the author of the paper. (Du Q, Thonberg H, Zhang HY, Wahlestedt C&Liang Z. Validating siRNA Using a Reporter Made from Synthetic DNA Oligonucleotides. Biochem Biophys Res Commun. 325:243-9, 2004; Du Q, Thonberg H, Wang J, Wahlestedt C&Liang Z.A Systematic Analysis of the Silencing Effects of an Active siRNA at All Single-nucleotide Mismatched Target Sites. Nucleic Acids Res. 33:1671-7, 2005; Li ZS, Qiao RP, Du Q, Yang ZJ, Zhang LR, Zhang PZ, Liang Z&Zhang LH. Studies on Aminoisonucleoside Modified siRNAs: Stability and Silencing Activity. Bioconjugate Chem.18:1017-24, 2007; Dahlgren C, Zha...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com