Method for detecting the methylation of colon-cancer-specific methylation marker genes for colon cancer diagnosis
By detecting the methylation status of colorectal cancer-specific methylation marker genes, the problem of difficulty in early diagnosis of cancer is solved, high-accuracy and high-sensitivity diagnosis of colorectal cancer is achieved, and a more effective method is provided. Treatment targets and monitoring tools.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
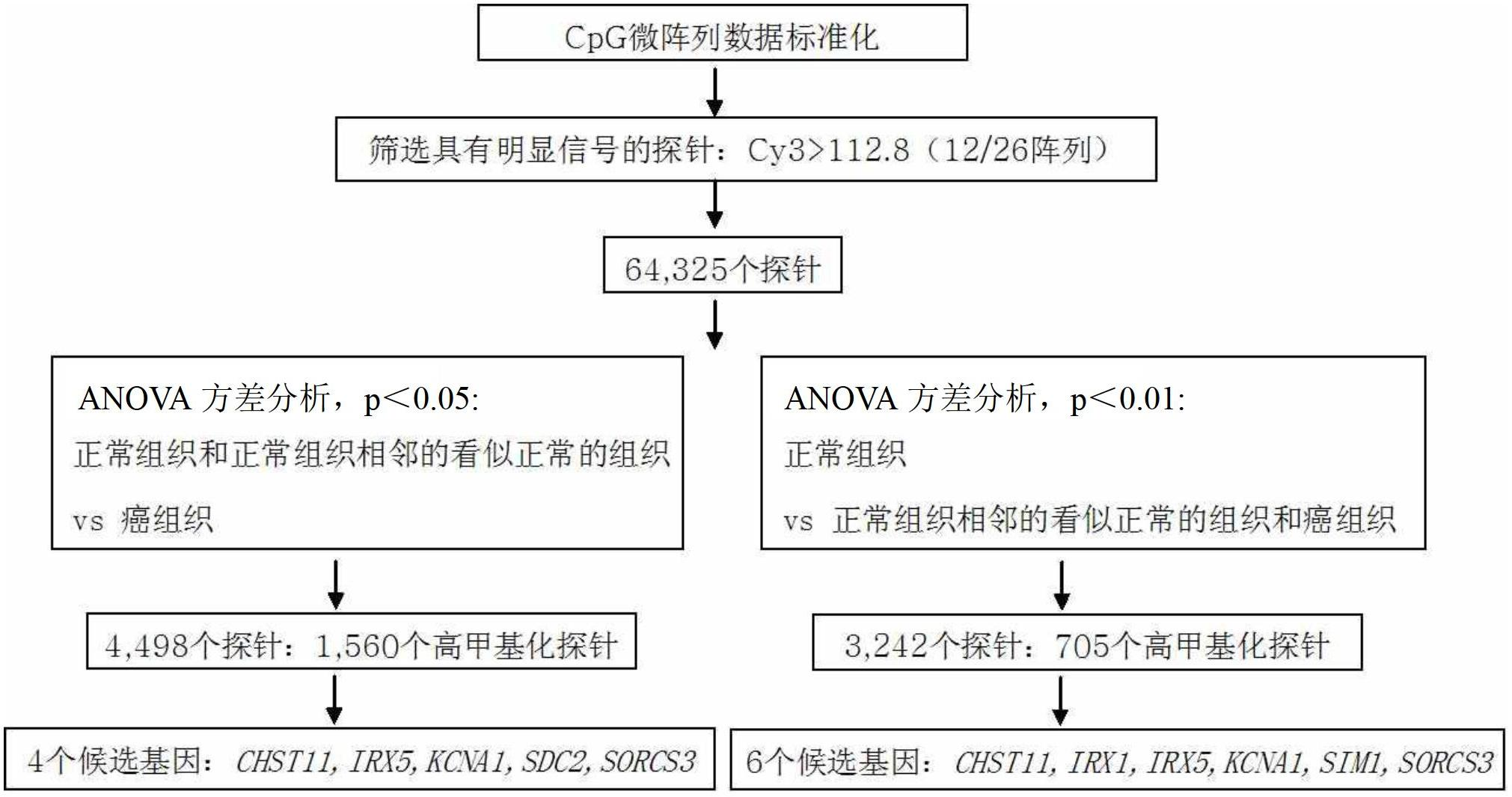
[0186] Example 1: Discovery of colorectal cancer-specific methylated genes
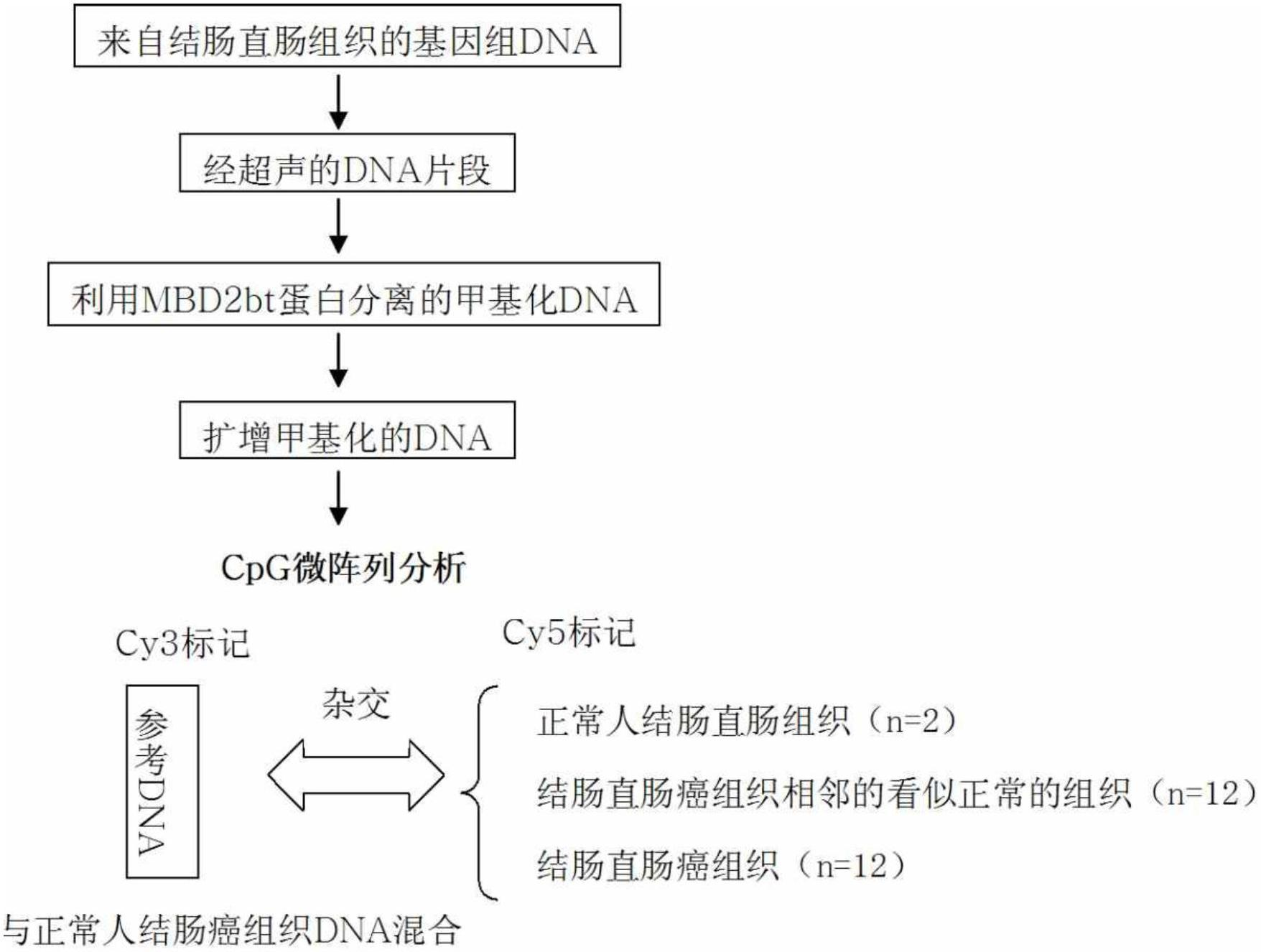
[0187] To screen for biomarkers specifically methylated in colorectal cancer, 500 ng each of genomic DNA from 2 normal individuals and from cancerous and adjacent normal tissues from 12 colorectal cancer patients were sonicated (Vibra Cell, SONICS), thereby constructing a genomic DNA fragment of about 200-300 bp.
[0188] To obtain only methylated DNA from the genomic DNA, a methyl binding domain (MBD) known to bind methylated DNA was used (Fraga et al., Nucleic Acid Res., 31:1765, 2003). Specifically, 2 μg of 6×His-tagged MBD2bt was pre-incubated with 500 ng of Escherichia coli JM110 (No. 2638, Biological Resource Center, Korea Research Institute of Bioscience & Biotechnology) genomic DNA. and then bound to Ni-NTA magnetic beads (Qiagen, USA). Genomic DNA isolated from normal people and colorectal cancer patients after sonication was 500ng each, in binding buffer (10mM Tris-HCl (pH 7.5), 50mM NaCl,...
Embodiment 2
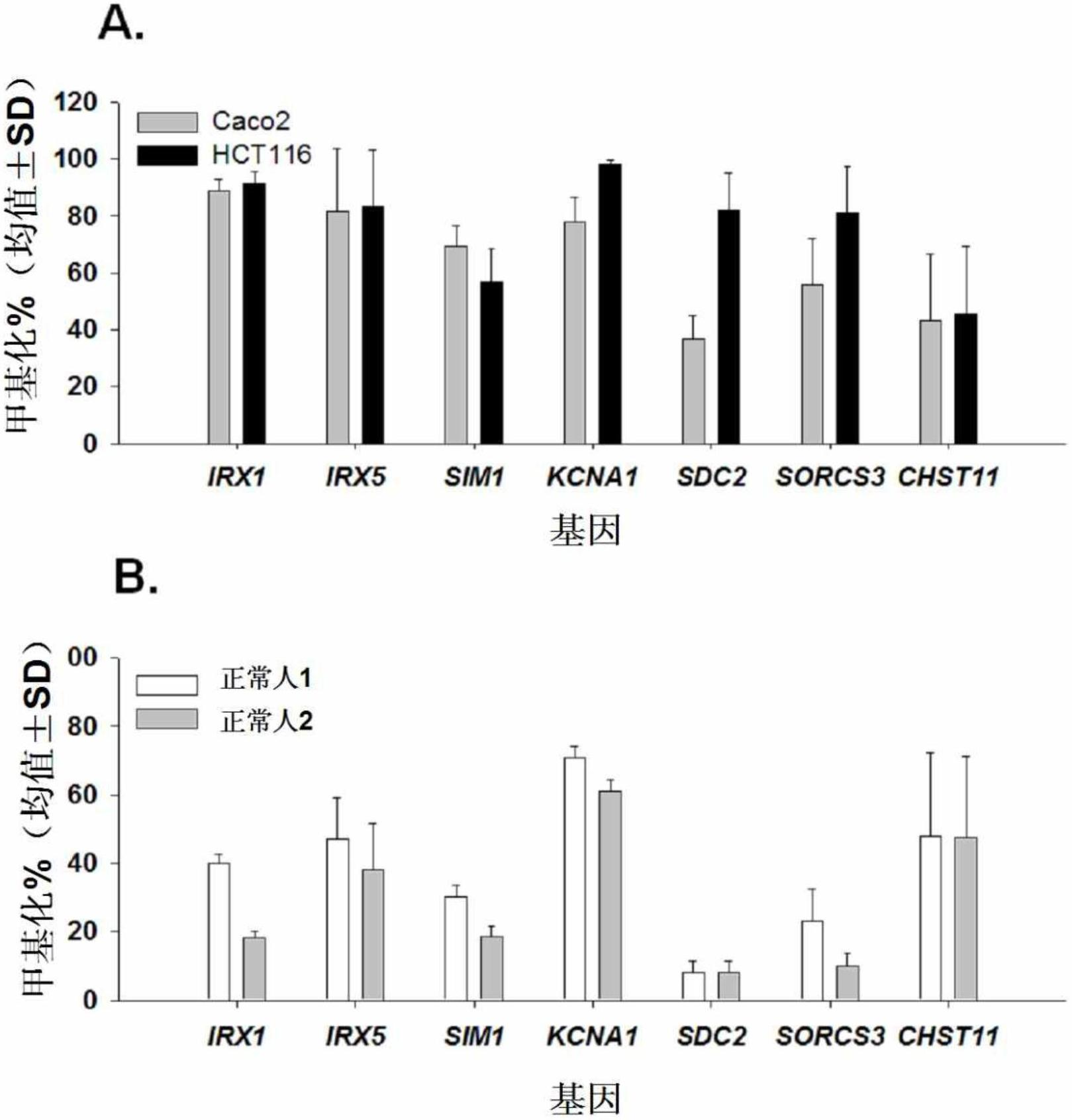
[0195] Example 2: Detection of Methylation of Biomarker Genes in Cancer Cell Lines
[0196] To further confirm the methylation status of the selected biomarker candidate genes in Example 1, pyrosequencing was performed on the promoter and intron regions of each gene.
[0197] To modify unmethylated cytosine to uracil with sulfite, total genomic DNA was isolated from colorectal cancer cell lines Caco-2 (KCLB No.30037.1) and HCT116 (KCLB No.10247) and analyzed by EZ DNA Methylation-Gold Standard Kit (Zymo Research, USA) treated 200 ng of the genomic DNA with sulfite. When the DNA is treated with sulfite, unmethylated cytosines are modified to uracils, while methylated cytosines remain unchanged. The sulfite-treated DNA was eluted in 20 μl sterile distilled water and subjected to pyrosequencing.
[0198] PCR primers and sequencing primers for pyrosequencing were designed for the 7 genes using the PSQ assay design program (Biotage, USA). The PCR primers and sequencing primers use...
Embodiment 3
[0209] Example 3: Detection of Methylation of Biomarker Candidate Genes in Normal Human Colorectal Tissue
[0210] In order for the seven biomarker candidate genes to be used as biomarkers for the diagnosis of colorectal cancer, these genes should show low methylation levels in colorectal tissues of normal people rather than patients, and should be found in colorectal cancer. Tissues exhibit high levels of methylation.
[0211] In order to confirm whether these genes meet these requirements, genomic DNA was isolated from two normal human colorectal tissues (Biochain) using the QIAamp DNA Mini Kit (QIAGEN, USA) and analyzed with the EZ DNA Methylation-Gold Standard Kit (Zymo Research, USA) treated 200ng of the isolated genomic DNA with sulfite. The treated DNA was eluted with 20 μl sterile distilled water and subjected to pyrosulfate sequencing.
[0212] 20 ng of the sulfite-treated genomic DNA was amplified by PCR. In the PCR amplification, PCR reaction solution (20 ng of s...
PUM
| Property | Measurement | Unit |
|---|---|---|
| frequency | aaaaa | aaaaa |
| percent by mass | aaaaa | aaaaa |
| percent by mass | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com