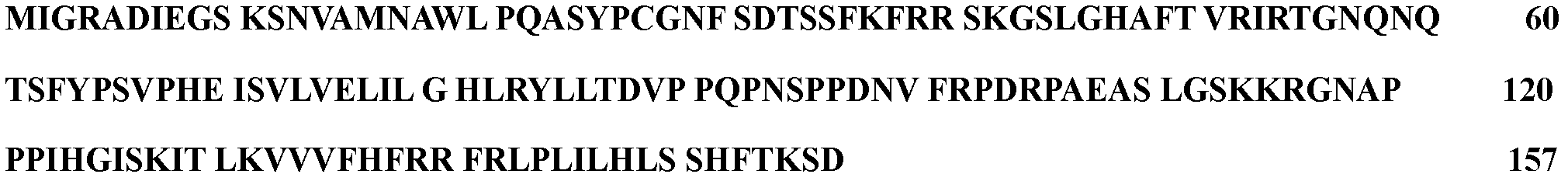Peanut senescence gene for regulating programmed cell death, coding sequence and application
A technology of programmed death and coding sequence, which is applied in the field of peanut aging gene AhSAG and its coding sequence and application, to achieve the effect of maintaining adaptability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Embodiment 1: RACE method obtains the full-length gene of AhSAG
[0071] 1. Synthesis of cDNA
[0072] According to different requirements, reverse transcription was performed to obtain different cDNAs, as shown in Table 3.
[0073] Synthesis of table 3AhSAG middle fragment, 3' end and 5' end cDNA
[0074] template
reverse transcription primer
purpose
99-1507400μmol / L Al treatment for 4d
OligoT18
normal template
99-1507400μmol / L Al treatment for 4d
F
3'race template
99-1507400μmol / L Al treatment for 4d
SAG 5′ reverse transcription
5'race template
[0075] 2. PCR amplification of the middle fragment, 3' end and 5' end cDNA fragment (50 μL system)
[0076] (1) Middle segment
[0077] reaction system:
[0078] The operation was performed on ice, and the specific operating conditions are shown in Table 3.
[0079] Table 3: Reaction system operating conditions
[0080] response system
v...
Embodiment 2
[0168] 实施例2、AhSAG cDNA正、反义植物表达载体的构建
[0169] 利用实施准备的材料和实施例1得到的AhSAG基因,将AhSAG基因插入载体pBI121-GFP(含GFP荧光蛋白)中GUS基因和35S启动子之间,并跟NOS终止子构成一个完整的表达框架。利用正义引物和反义引物通过PCR扩增基因AhSAG的开放阅读框(ORF),纯化回收后,克隆到空载体pTG19-T(上海生工),获得正义质粒pTG19-AhSAG和反义质粒pTG19-anti-AhSAG。将含有AhSAG的pTG19-AhSAG和pTG19-anti-AhSAG cDNA的克隆载体用BamH I和XbaI双酶切,分别回收小片段,定向连接到经BamH I和XbaI双酶切的载体pBI121-GFP上,连接产物转化E.coli DH5α感受态细胞、碱法提取质粒。正义载体命名为pBI121-GFP-AhSAG;反义载体命名为pBI121-GFP-anti-AhSAG。
[0170] 1、AhSAG基因编码区cDNA序列的克隆
[0171] (1)RT-PCR(50μL体系)
[0172] reaction system:
[0173] 采用冰上操作,具体操作条件见表13。
[0174] 表13:反应体系操作条件
[0175] response system
volume
10×Buffer (including Mg 2+ )
5μL
dNTP (2.5mM)
4μL
P1: SAG sense / antisense upstream (10 μM)
2μL
P2: downstream of SAG sense / antisense (10 μM)
2μL
OligoT18 reverse transcription product stock solution
1μL
Ex-Taq
0.25 μL
wxya 2 o
35.75μL
total
50μL
[0176] Reaction conditions:
...
Embodiment 3
[0206] 实施例3:AhSAG基因在不同品种和不同铝胁迫下表达量的变化
[0207] 选用Actin基因做内参,提取8个材料的RNA,以oligoT18为引物进行反转录,得到8个材料的cDNA,分别记为99-0、99-20、99-100、99-400、中2-0、中2-20、中2-100和中2-400,进行RT-PCR(Real-time PCR)实时。
[0208] (1)反应体系:
[0209] 采用冰上操作,具体操作条件见表17。
[0210] 表17:反应体系操作条件
[0211] reaction system
volume
Hotstart Fluo-PCR Mix
12.5μL
P1 (10μM)
1μL
P2 (10μM)
1μL
cDNA (100mM)
0.5μL
wxya 2 o
10μL
total
25 μL
[0212] (2)反应条件:
[0213] 具体反应操作条件见表18。
[0214] 表18:反应操作条件
[0215]
[0216]
[0217] 根据2 -△△t 法计算相对表达量。
[0218] 结果:中花2号和99-1507的表达量的趋势基本一致,只是在低浓度下,中花2号的表达量有所下降,有可能是低浓度的Al促进了植物的生长,导致衰老基因的表达下调。但当在中、高浓度处理时,衰老基因的高丰度表达。由此,可以认为AhSAG基因属于衰老诱导增强型基因,中、高浓度下诱导AhSAG等衰老基因超表达,促进植物早衰。
[0219] From the expression of AhSAG gene and the relative expression of AhSAG gene between Zhonghua 2 and 99-1507, the expression of the aluminum-sensitive variety Zhonghua 2 suddenly increased under the treatment of medium and hi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com