Method for quantitatively detecting protein through quantum dot resonant scattering
A quantitative detection and quantum dot technology, which is applied in the field of protein detection, can solve the problem of unquantified detection of proteins, etc., and achieve the effect of high sensitivity and good stability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] 1) Preparation of ZnS / ZnO quantum dot heterostructure: First, 2mmol Zn(Ac) 2 and 1.4 mmol CH 3 CSNH 2 Add 20ml of 1,4-butanediol, and ultrasonically disperse until a transparent precursor solution is formed; then transfer the obtained solution to a 30mL hydrothermal reaction kettle (lined with polytetrafluoroethylene), and react at 200°C for 18h; After the thermal reaction kettle was naturally cooled to room temperature, 10-20ml of distilled water was added to the obtained product to mix, centrifuged at 8000rpm, the obtained precipitate was washed with ethanol three times to remove impurities, and dried at 60°C for 5 hours to obtain the ZnS / ZnO quantum dot heterostructure. The diameter is 3~4nm.
[0038] 2) Silicon-coating step: Dissolve 0.1mmol ZnS / ZnO quantum dots in 40mL ethanol, add 0.5mL 28wt% ammonia water and mix well, then slowly add 0.1mL TEOS under continuous stirring, and continue stirring for 12 hours. Centrifuge to get precipitate, wash twice with distil...
Embodiment 2
[0041] Embodiment 2 takes the detection of bovine serum albumin (BSA) as an example
[0042] (1) Preparation of quantum dot solution and protein solution
[0043] The original concentration of quantum dots 1 to 4 is 5×10 -5 mol / L, diluted 60 times with double distilled water, and ultrasonicated for 15 minutes before use to make it evenly suspended.
[0044] BSA formulated at a concentration of 10 -4 mol / L, spare. BSA was purchased from Beijing Suolaibao Biotechnology Company.
[0045] (2) Detection and drawing of standard curve
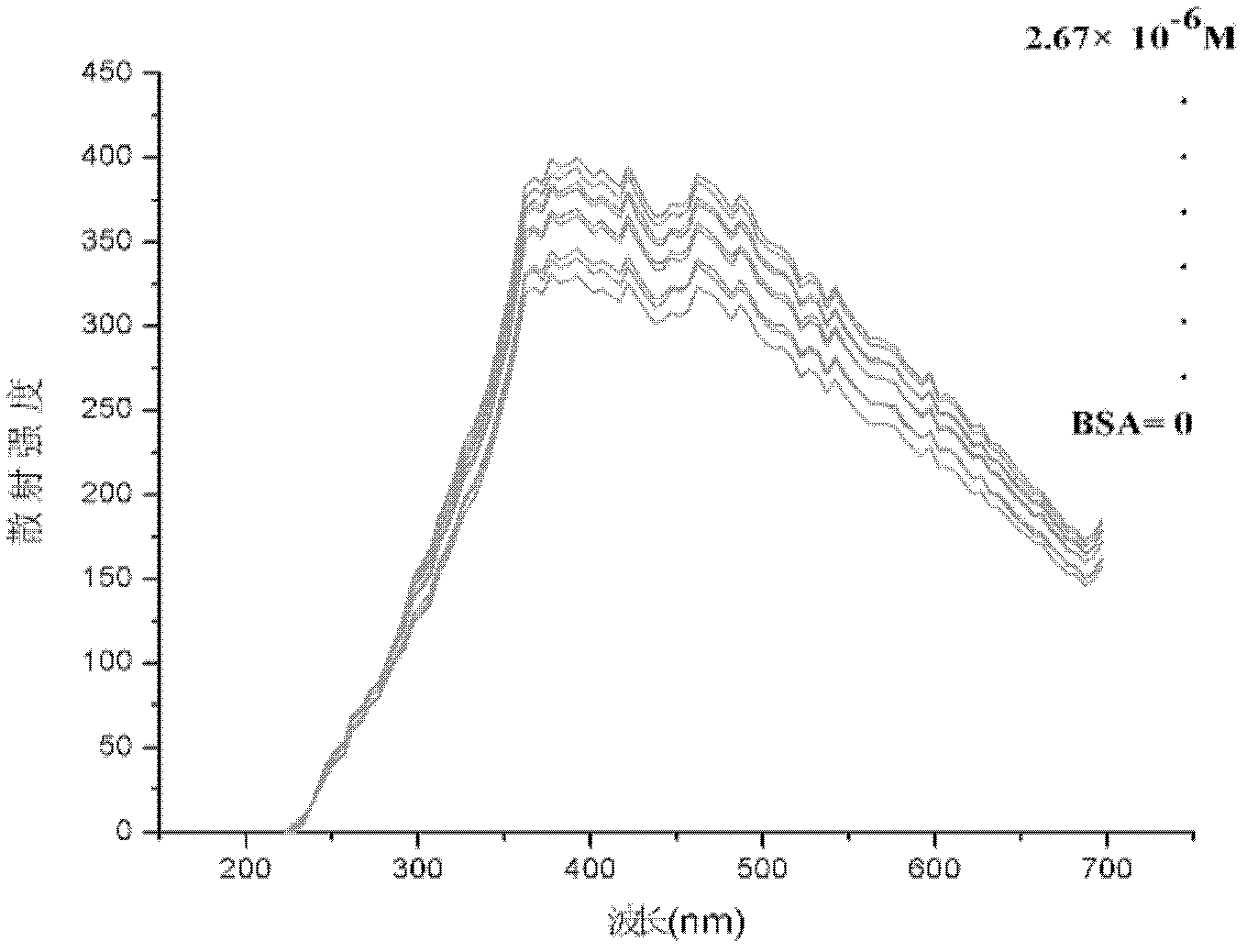
[0046] After the instrument is preheated stably, take 3mL of pure quantum dots of one of the particle sizes in a cuvette, and perform simultaneous fluorescence detection at 250-750nm, the emission grating and scattering grating are both 5nm, the voltage is 400V, and the scattering intensity is f 0 .
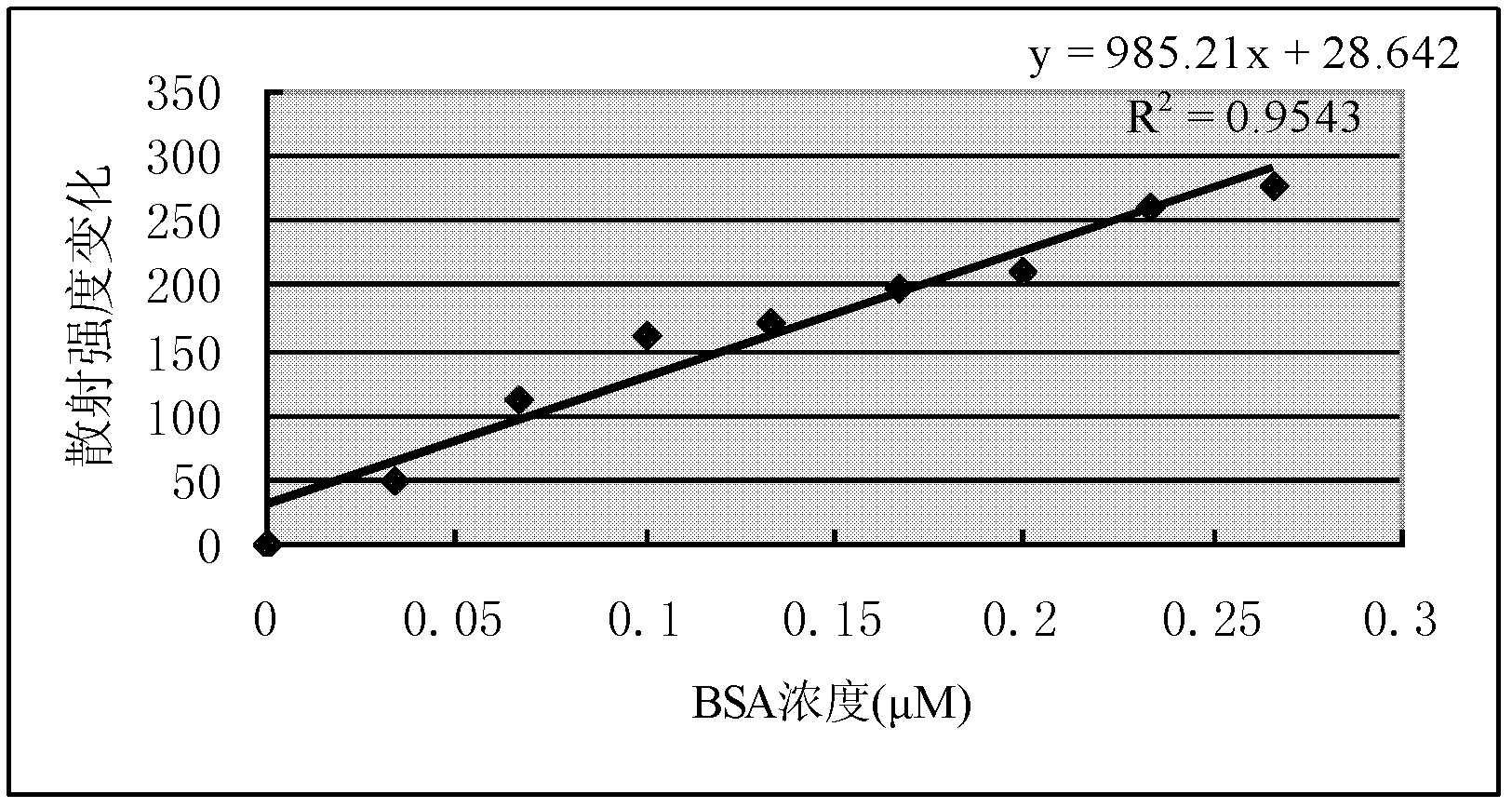
[0047] Then add 10, 20, 30, 40, 50, 60, 70, 80, 90 and 100 μL respectively to a concentration of 10 -4 mol / L BSA solution; each step is tested f...
Embodiment 3
[0049] Example 3 Detection of gamma-globulin
[0050] (1) Preparation of quantum dot solution and protein solution
[0051] The original concentration of quantum dots 1 to 4 is 5×10 -5 mol / L, diluted 60 times with double distilled water, and ultrasonicated for 15 minutes before use to make it evenly suspended.
[0052] The concentration of γ-globulin is 10 -4 mol / L, spare. γ-globulin was purchased from Beijing Suolaibao Biotechnology Company.
[0053] (2) Detection and drawing of standard curve
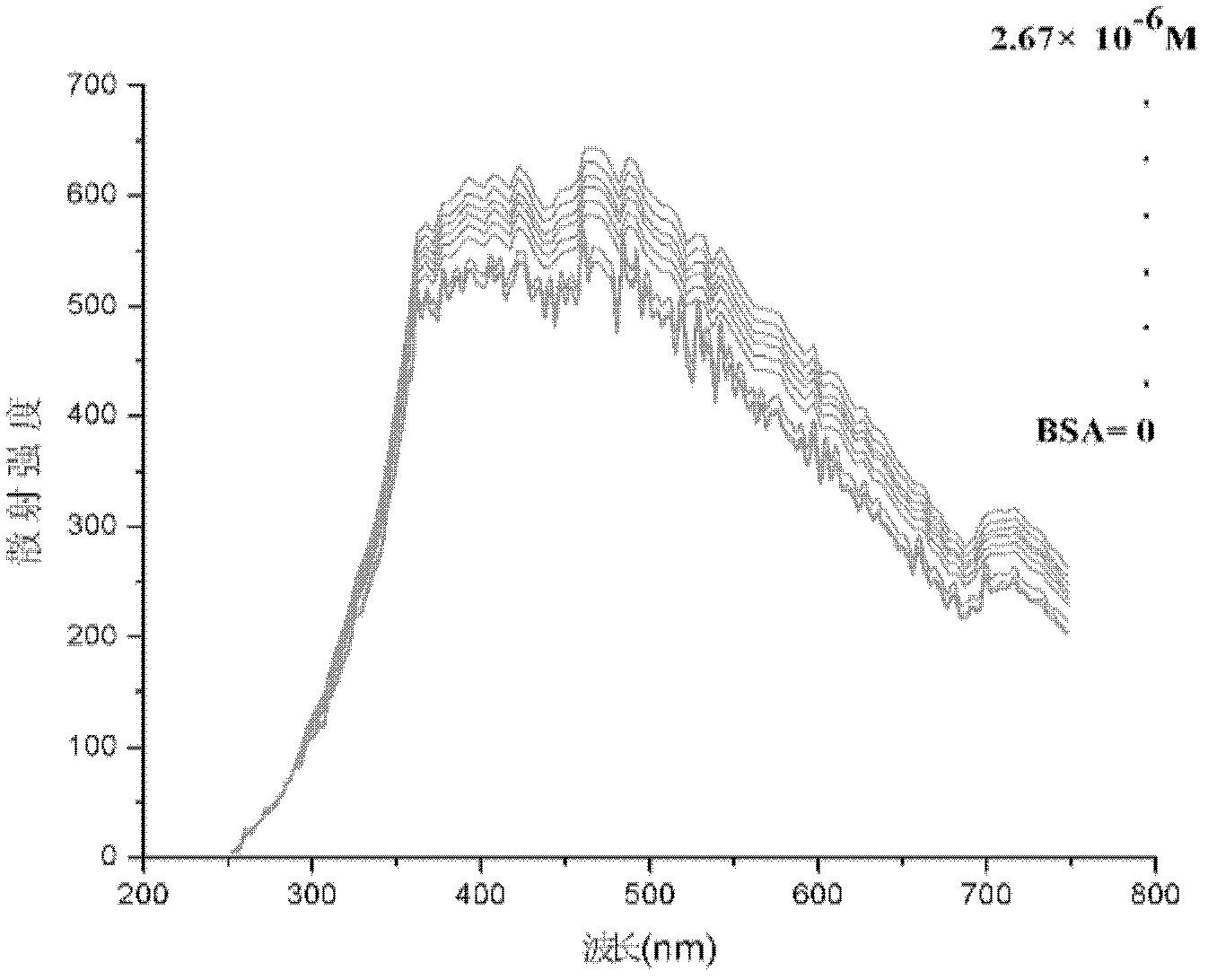
[0054] After the instrument is preheated stably, take 3mL of pure quantum dots of one of the particle sizes in a cuvette, and perform simultaneous fluorescence detection at 250-750nm, the emission grating and scattering grating are both 5nm, the voltage is 400V, and the scattering intensity is f 0 . Then add 10, 20, 30, 40, 50, 60, 70 and 80 μL of 10 -4 mol / L of γ-globulin, each step is tested for zeroing, and the scattering intensity is F n . It is found that the maximum sc...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com