Saccharomyces cerevisiae strain and method for selecting saccharomyces cerevisiae strains expressing activated xylose translocator
A technology of Saccharomyces cerevisiae strain and xylose transporter, which is applied in microorganism-based methods, biochemical equipment and methods, fungi, etc., can solve the problems of large sedimentation coefficient, difficulty in molecular entry and exit, and different xylose transport mechanisms. Achieve the effect of avoiding false positives, simple operation steps, and less demanding operators and equipment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
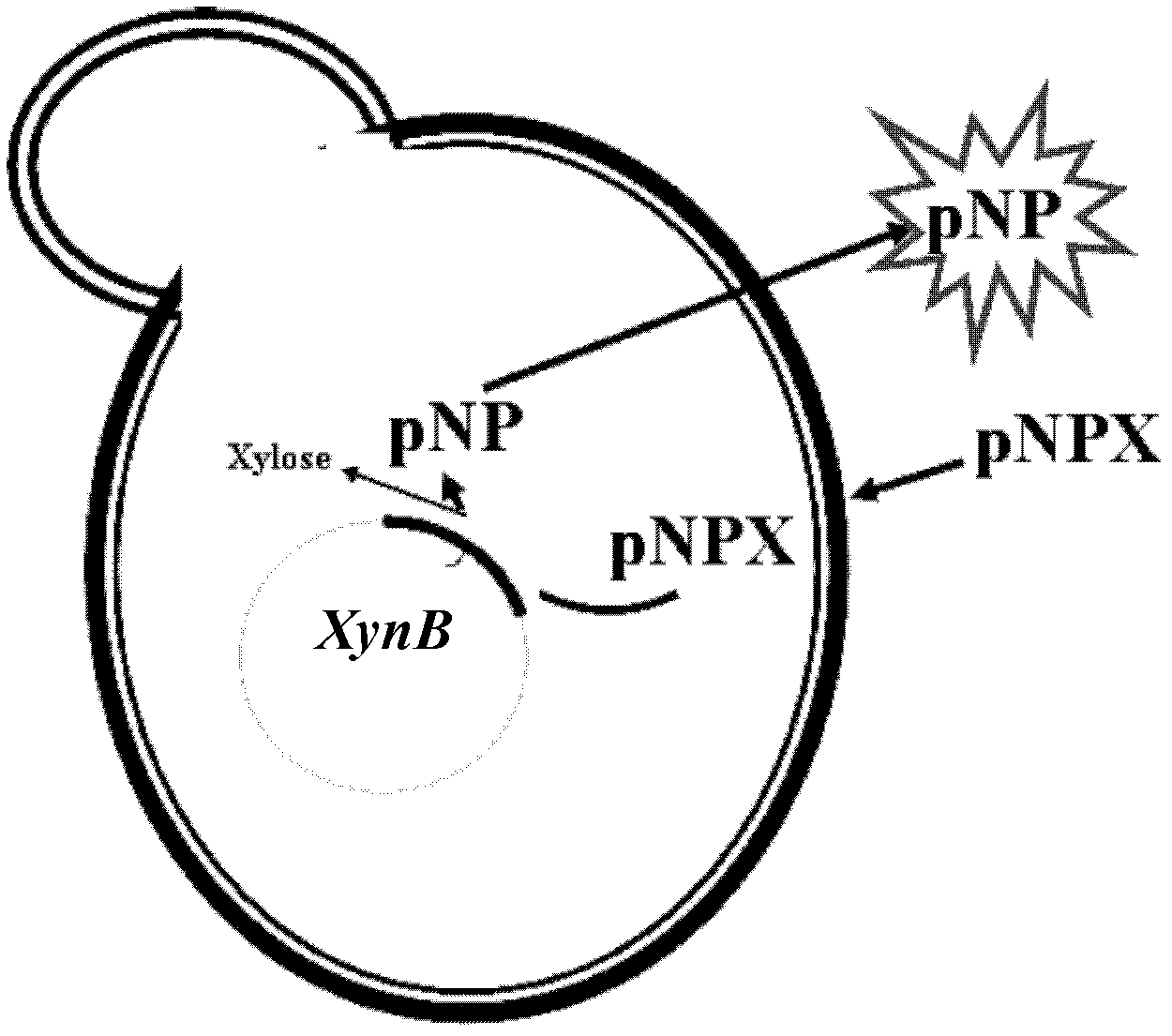
[0043] Example 1: Construction of strain BSW2ABP expressing xylosidase and assay of xylosidase enzyme activity
[0044] Methods as below:
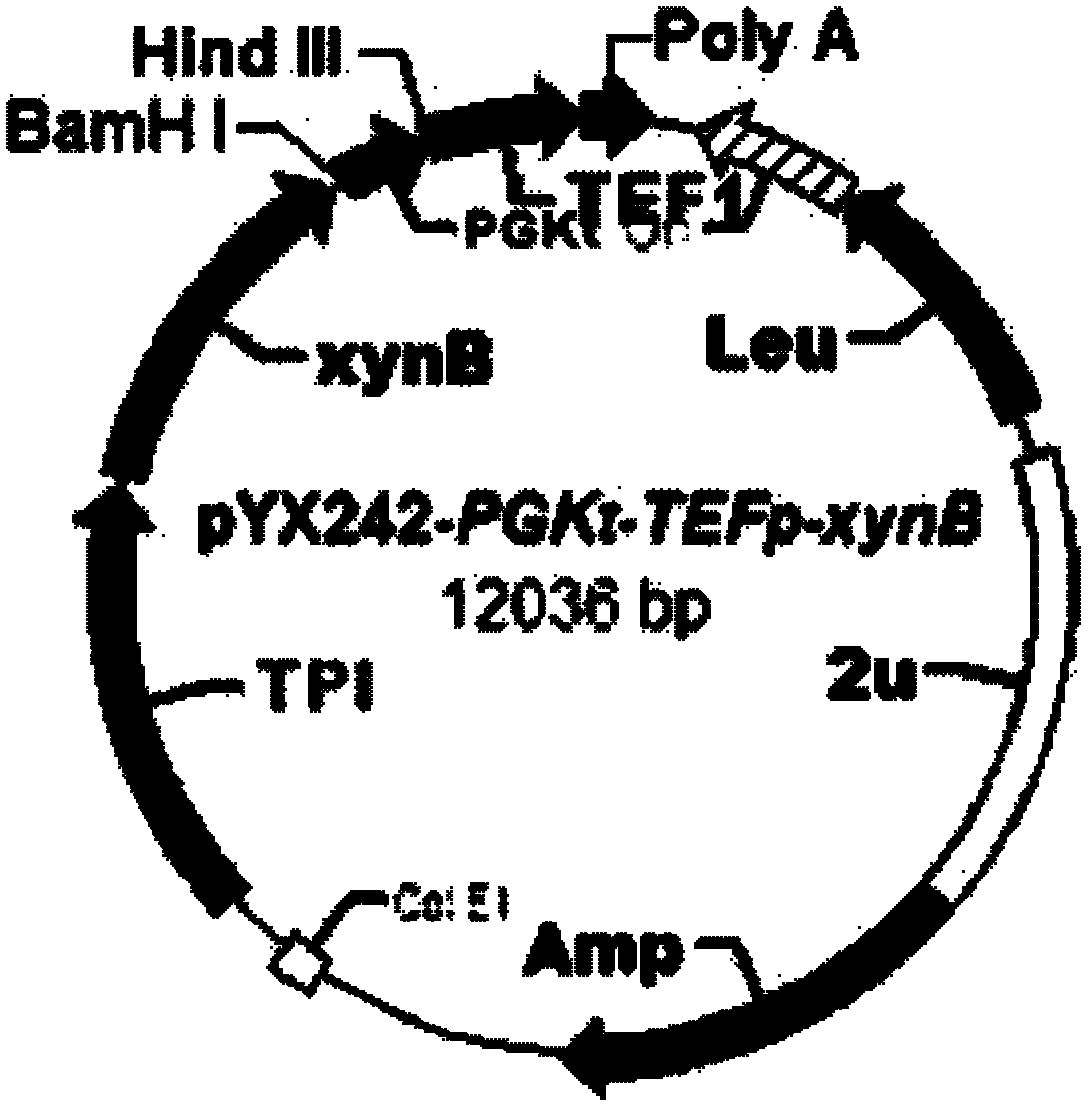
[0045] (1) Strain selection: select the constructed laboratory strain BSW2AB (CEN.PK102-3A derivative, MATa, leu2-3, ura3-52, pYX242-PGKt-TEFp TPIp-xynB) with intracellular heterologous expression of xylosidase -PGKt) is used as the recipient strain, and the strain is classified as Saccharomyces cerevisiae, and has been deposited in the "General Microorganism Center of China Committee for the Collection of Microbial Cultures" on November 17, 2011, with a preservation number of CGMCC No.5465 . Its mycological characteristics are: CEN.PK102-3A derivative, MATa, leu2-3, ura3-52, pYX242-PGKt-TEFpTPIp-xynB-PGKt, the yeast strain is a unicellular eukaryotic microorganism, oval, immobile, solid Or under the culture condition of the fully synthetic auxotrophic culture medium (SC-Leu, this culture medium is existing in the prior art) of liquid, b...
Embodiment 2
[0054] Example 2: Determination of the relationship between incubation time and transport rate measurements
[0055] Methods as below:
[0056] (1) Select the bacterium liquid cultivated for 15 hours in the embodiment 1 step (4), centrifuge (3000r min -1 , 5min) resuspended with 50mmol / L PBS solution of pH 6.5 after removing the original culture medium;
[0057] (2) Determination of the relationship between the incubation time and the measured value of the transport rate: Take an appropriate amount of the complete cell suspension in step (1) with a 96-well plate or EP tube, add pNPX with a final concentration of 6 mmol / L, and then place it at 30 ℃, 200r·min -1 In a shaker, incubate for 10min, 20min, 30min, 40min, 50min and 60min respectively, centrifuge to take 100μl of the supernatant and add it to a 96-well plate, then add an equal volume of 0.4mol / L Na 2 CO 3 Aqueous solution, after mixing, use a microplate reader at OD 405nm The light absorbance of p-nitrophenol (pNP)...
Embodiment 3
[0058] Example 3: Determination of the extracellular and extracellular content of pNP under the condition of complete cell determination
[0059] Methods as below:
[0060] (1) Select the bacterium liquid cultivated for 15 hours in the embodiment 1 step (4), centrifuge (3000r min -1 , 5min) resuspended with 50mmol / L PBS solution of pH 6.5 after removing the original culture medium;
[0061] (2) Determination of the extracellular content of pNP in the state of intact cells: use a 96-well plate or EP tube to take an appropriate amount of the complete cell suspension in step (1), add pNPX with a final concentration of 6mmol / L, and then place it at 30 ℃, 200r·min -1 Incubate on a shaker for 10min and 30min respectively; centrifuge to take 100μl of the supernatant and add it to a 96-well plate, then add an equal volume of 0.4mol / L Na 2 CO 3 Aqueous solution, after mixing, use a microplate reader at OD 405nm The light absorbance of p-nitrophenol (pNP) produced after incubation ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com