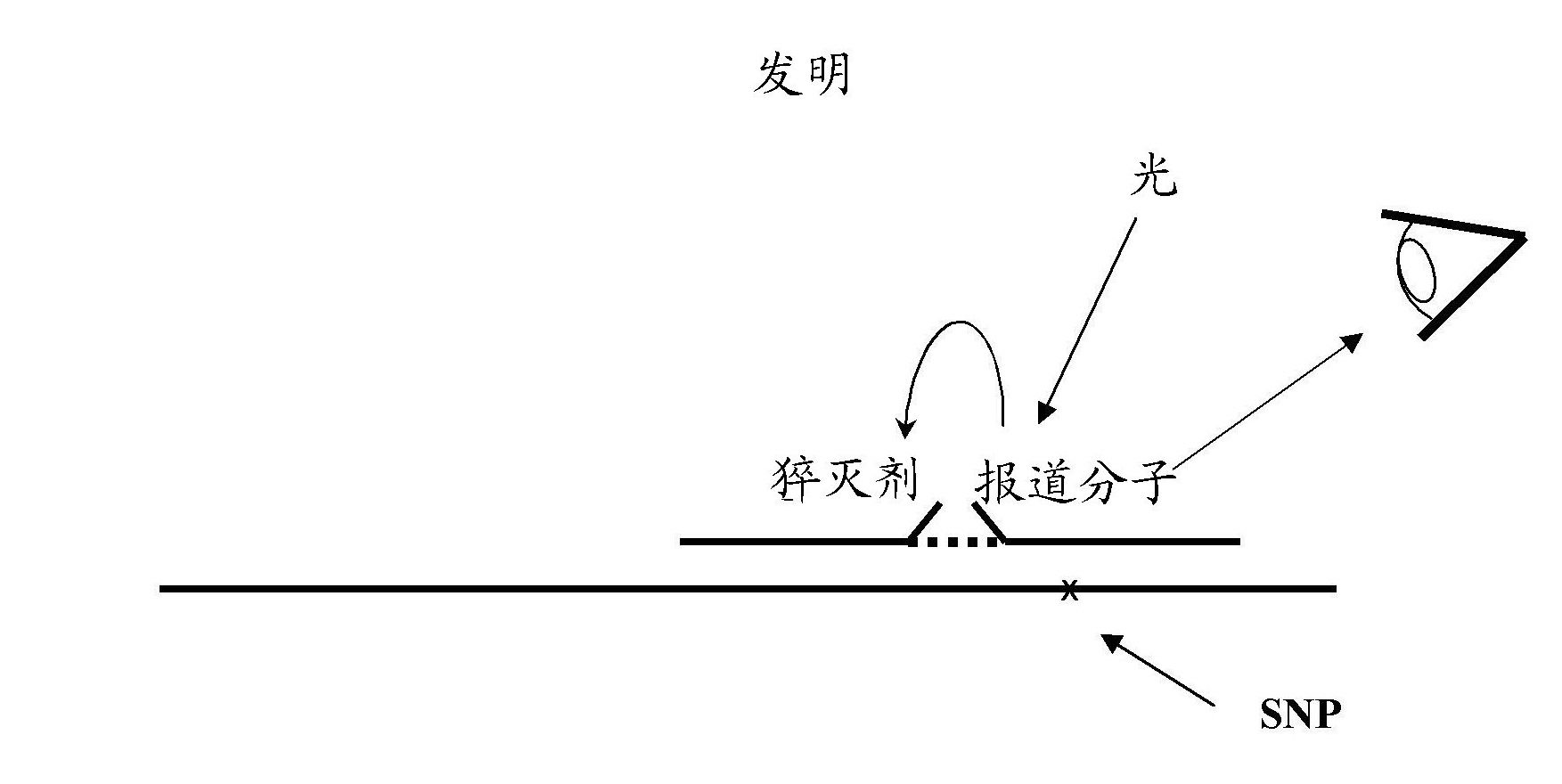
Method for detecting a target nucleic acid in a sample
A technology for target nucleic acid and target detection, applied in the field of target nucleic acid detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
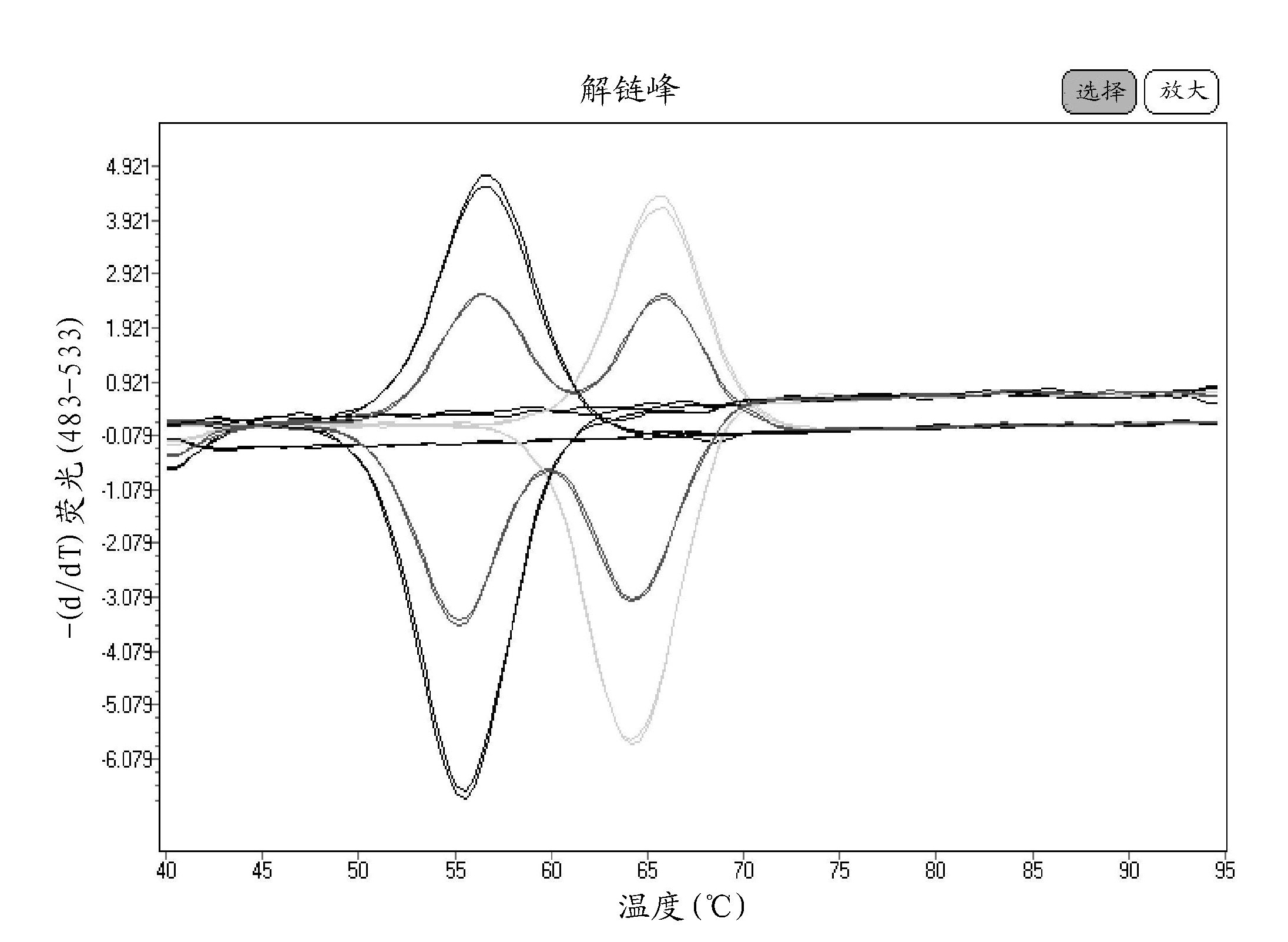
[0074] In these examples, the method of the present invention was used to amplify the region of the human Factor V gene containing the Leiden mutation site and cloned into a plasmid vector. In the current example, the asymmetric PCR sample master mix (100 uL) consisted of the following components: 5% glycerol; 50 mM Tricine, pH 8.3; 25 mM potassium acetate; 200 μM dATP, 200 μM dGTP, 200 μM dCTP, 400 μM dUTP; 0.7 μM upstream (excess) primer; 0.1 μM downstream (limiting) primer; 0.4 μM each probe; 0.04 U / μL uracil-N-glycosylase; 0.4 U / μL ΔZO5 DNA polymerase; and 4 mM magnesium acetate.
[0075] Master mixes are used to amplify Factor V wild-type, mutant, and mixed plasmid DNA targets. Excess primer was present at 7 times the limiting primer concentration to ensure an excess of single stranded amplicon bound to the hybridization probe. Amplification and melting were performed on a Roche LightCycler 480.
[0076] The thermal cycle profile used in this example was: 50°C for 5 min...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com