MLCR probe, two-step reaction mode and suspension chip detection capture probe
A probe and reaction technology, applied in the field of MLCR probe, two-step reaction mode and capture probe for suspension chip detection, can solve the problem of low detection sensitivity and achieve the effect of improving specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
[0078] Specific implementation mode 1: Triple gene detection
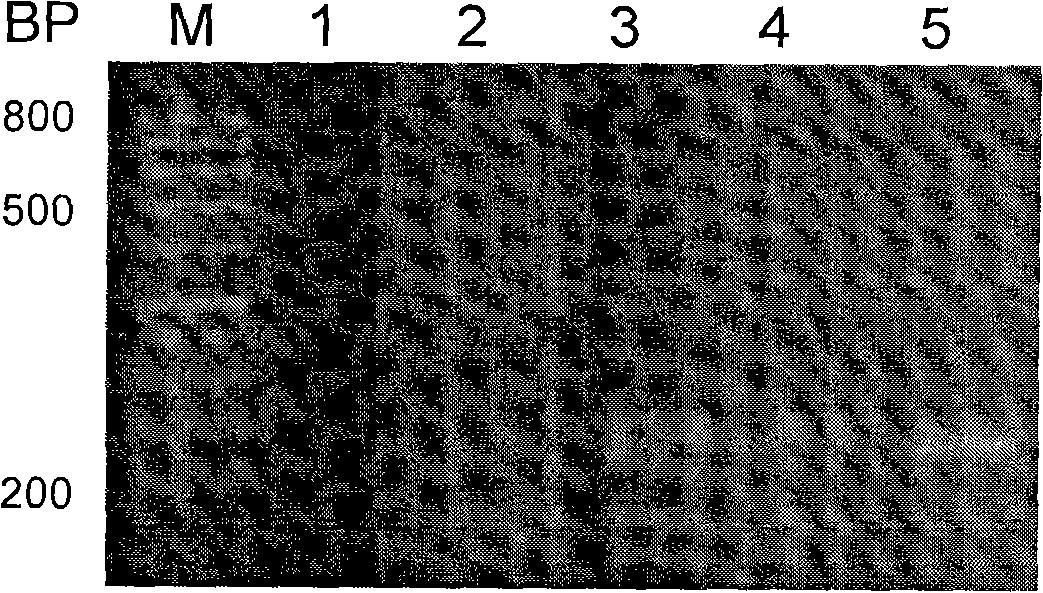
[0079] In order to visually test the reliability of MLCR-PCR detection technology, gel electrophoresis separation method was used to visually analyze MLCR-PCR reaction products. For this reason, the following three-site probes were synthesized by chemical method, and triple gene detection was performed first.
[0080] 1. Synthetic oligonucleotide sequence:
[0081] MLCR probe sequence
[0082]
[0083] Primers for PCR reactions
[0084] Primer-F: GGGTTCCCTAAGGGTTGGA;
[0085] Primer-R: GCGCCAGCAAGATCCAATCTAGA.
[0086] 2. MLCR reaction
[0087] MLCR reaction system:
[0088] Sample genomic template DNA: 1μl (20ng / μl)
[0089] Mixed probes: 2 μl (10 fmol / μl per probe)
[0090] 10x buffer: 1 μl
[0091] Ampligase: 0.2 Unites
[0092] wxya 2 O: 5.9 μl
[0093]
[0094] Total reaction volume: 10 μl
[0095] MLCR reaction conditions:
[0096] 2 cycles: 94°C, 1....
specific Embodiment approach 2
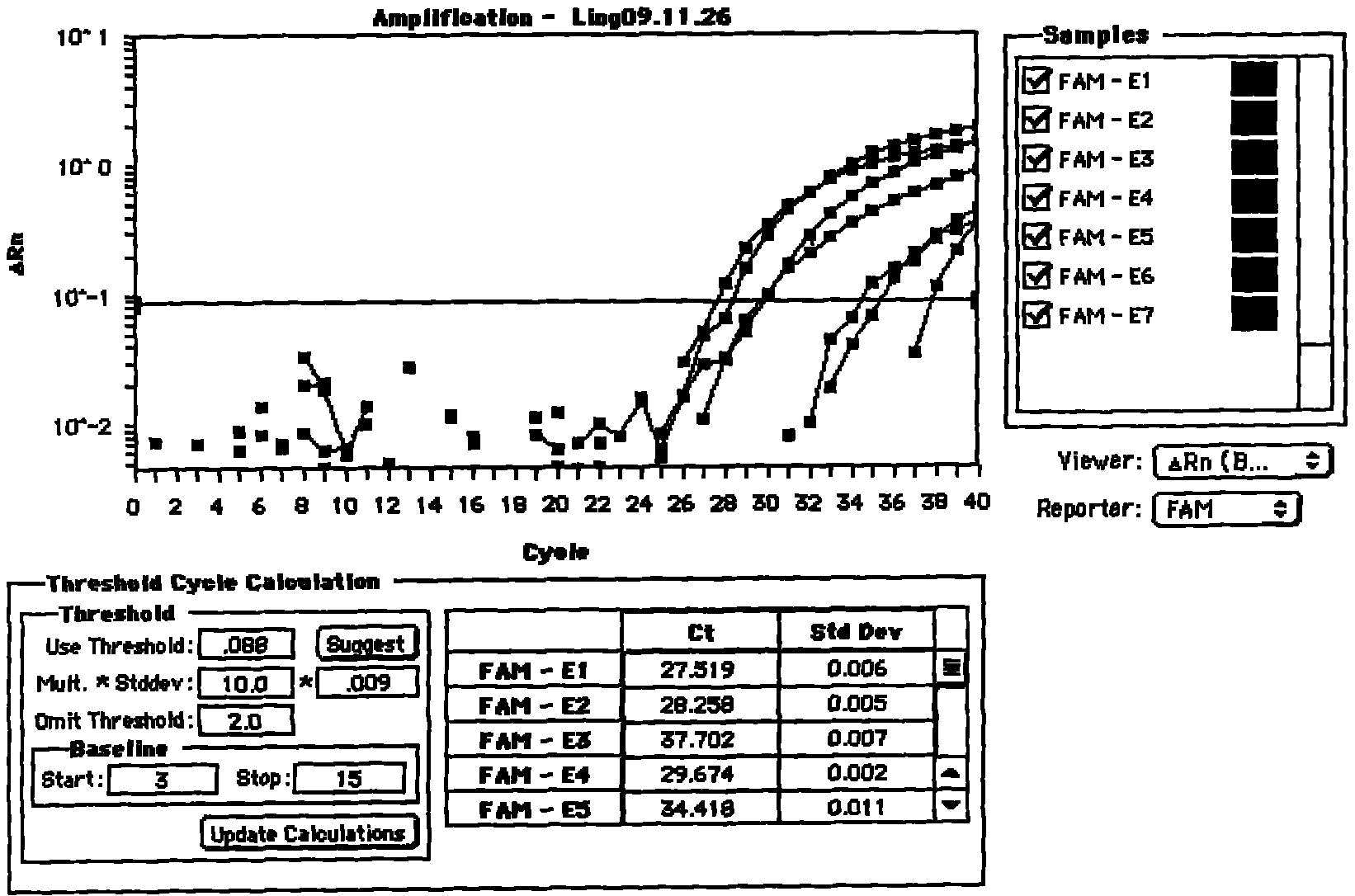
[0118] Specific implementation mode two: Seven-fold gene detection
[0119] 1. Synthetic Oligonucleotide Sequence
[0120] LCR probe sequence
[0121]
[0122]
[0123]
[0124] Primers for PCR reactions
[0125] Primer-F: GGGTTCCCTAAGGGTTGGA;
[0126] Primer-R: Bio-GCGCCAGCAAGATCCAATCTAGA (5'biotinylated).
[0127] Bio-Plex Capture Probes
[0128] capture probe name
Sequence (5'-3')
modify
CapNOS
cttgtccaagatctattcaggtgcac
26bp
5' end linking 5' amino repair
Decorated C12 arm
Cap35S
cagtggtcccaaagatggacccccac
26bp
5' end linking 5' amino repair
Decorated C12 arm
CapLec
aagttacaactcaataaggttgacg
25bp
5' end linking 5' amino repair
Decorated C12 arm
Cap RRS
ctccgcacaggtgaagtccgccgtgc
26bp
5' end linking 5' amino repair
Decorated C12 arm
CapGos
cagtgtggttggtttcttcggacgc
25bp
5'...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com