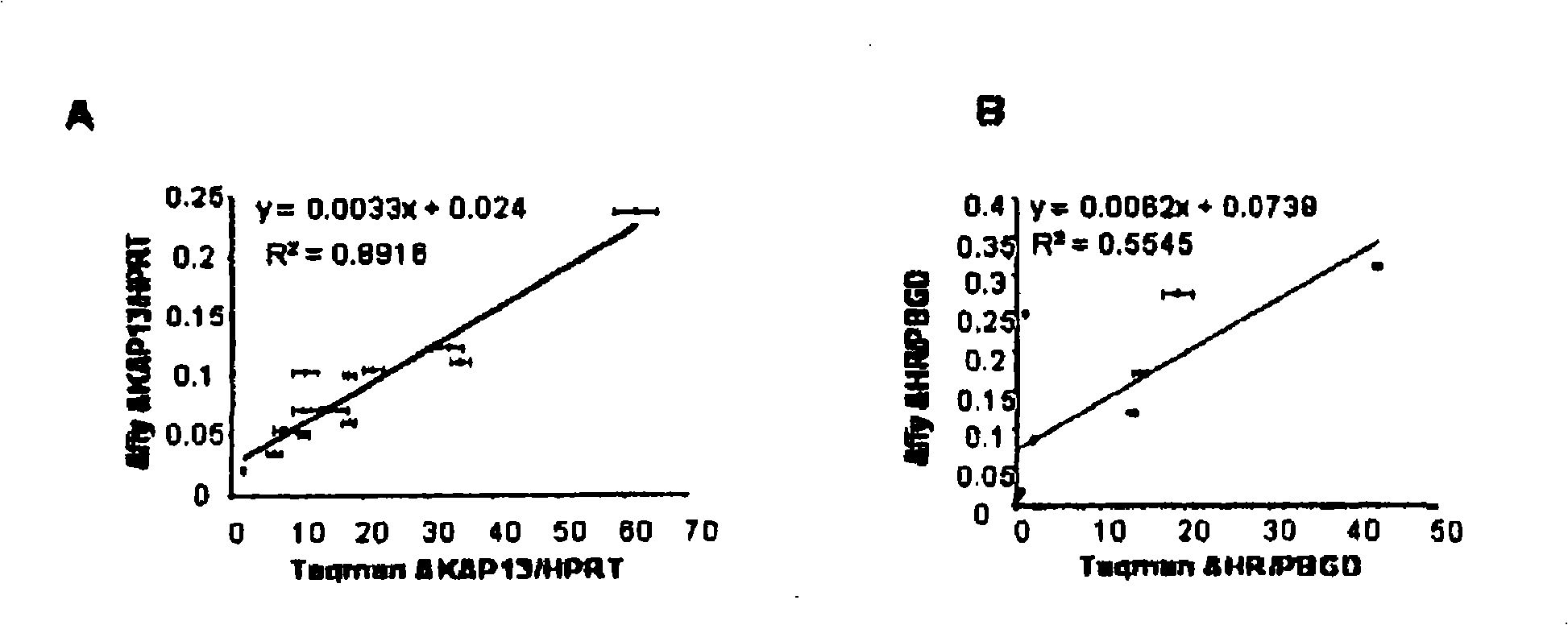Methods for assessing patients with acute myeloid leukemia
A technology for myeloid leukemia and patients, applied in the fields of biochemical equipment and methods, organic chemistry, microbial determination/examination, etc., can solve problems such as complex inhibitory effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0112] Definition of clinical valuation and response
[0113] The current study is part of a publicly labelled, multicenter, non-comparative Phase 2 clinical study, in which patients with relapsed or refractory AML (Harousseau et al. (2003)) are treated with tipifarnib, for For the first 21 consecutive days of each 28-day cycle, the initial oral dose is 600 mg twice a day. The patients were divided into two groups: those with relapsed AML and those with refractory AML. A total of 252 patients (135 relapsed and 117 refractory) were treated. Eighty patients were selected to provide bone marrow samples for RNA microarray analysis, and patient consent was separately obtained for this. The overall response rate in this study was relatively low.
[0114] Therefore, for the purpose of gene expression profiling, the response to tipifarnib is defined as the patient having an objective response (complete remission [CR], complete remission with incomplete platelet recovery [CRp] or partial...
Embodiment 2
[0143] Identify genes that are differentially expressed between responders and non-responders
[0144] Second, we used gene expression data to conduct a supervised analysis to identify genes that are differentially expressed between all responders and at least 40% of non-responders. These criteria can be selected to identify genes with the highest possible level of sensitivity that can predict the response to tipifarnib. From the 11,723 genes, a total of 19 genes that can classify responders and non-responders and give a significant P value in the t-test were identified (see Table 3 and Table 10 for more details). These genes include those involved in signal transduction, apoptosis, cell proliferation, tumorigenesis and possibly FTI biology (ARHH, AKAP13, IL3RA).
[0145] Table 3 19 main genes predicting response to tipifarnib and analysis results
[0146]
[0147] Real-time RT-PCR confirmation of genetic markers
[0148] To verify the microarray gene expression data, TaqMan...
Embodiment 3
[0165] Analysis of AML prognostic gene markers
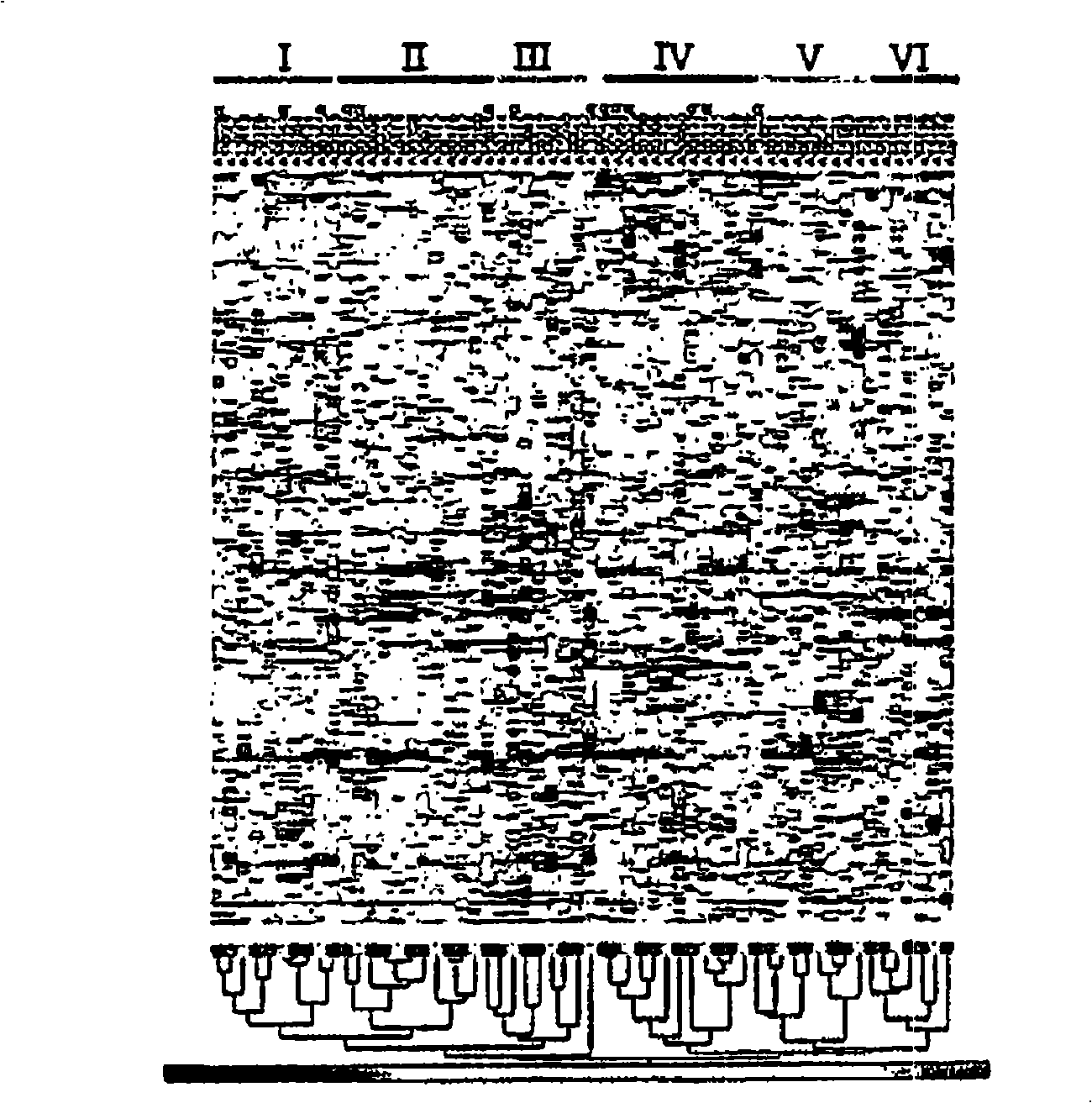
[0166] The 3 gene markers can predict prognosis independently of the type of drug treatment. To confirm this, we first evaluated gene expression markers recently identified in newly diagnosed AML patients treated with conventional chemotherapy. Bullinger et al. (2004). A cDNA array was used to define this marker, so we first matched these genes with the probes present on the Affymetrix gene chip. Among the 133 predictive genes identified by Bullinger et al., 167 probe sets (corresponding to 103 unique genes) matched the Affymetrix U133A chip. The 3 genes identified in our current analysis are not in the 133 gene list of Bullinger et al. SEQID NOs: shown in Table 4. Using these 167 probe groups, two main patient groups were defined by hierarchical clustering (Figure 8A). Kaplan-Meier analysis showed that these clusters were clearly stratified into patients with good and poor prognosis (Figure 8B, p=0.000003). Therefore, our data co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com