Method for extracting and purifying target nucleic acid and use thereof
An extraction method and technology of target nucleic acid, applied in the field of bioengineering, can solve the problems of inappropriateness and high sensitivity requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
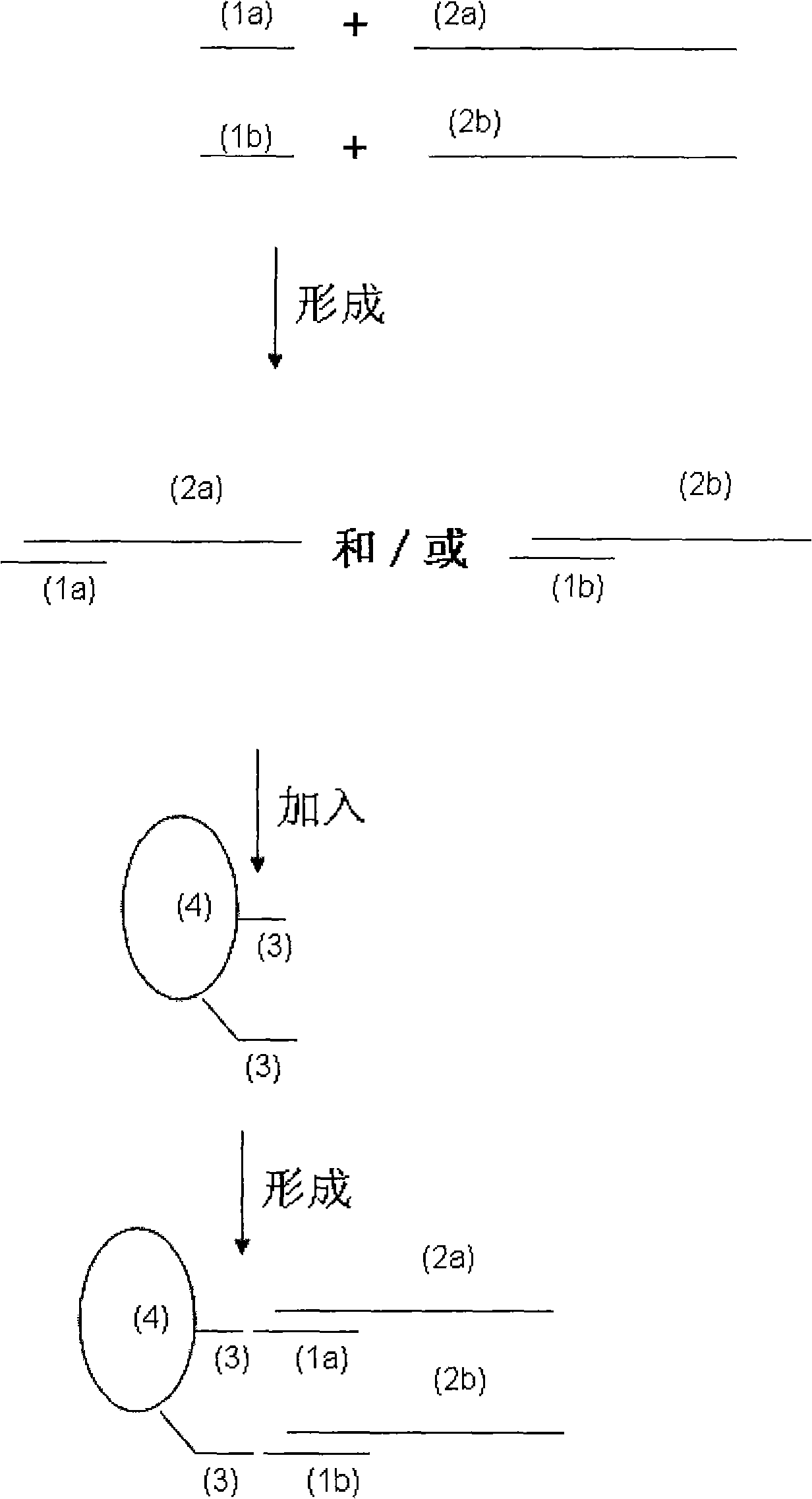
[0065] Embodiment 1, utilize 2 '-deoxy oligonucleic acid (DNA) capture probe and 2 '-oxygen-methylated oligonucleic acid (2'-OME-RNA) capture probe to capture the comparison of HCV RNA target efficiency
[0066] Taking HCV RNA (sequence 2 in the sequence listing) as an example, using Biotin (biotin) as the marker B and Strepavidin (streptavidin) as the marker B specific binder A to detect the 2'-deoxy oligonucleotide capture probe and 2'-oxygen-methylated oligonucleotide capture probes to the capture efficiency of RNA target nucleic acid, the specific method comprises the following steps:
[0067] 1. Reagent preparation
[0068] 1. Lysis solution (A) with 2'-deoxyoligonucleotide (DNA) capture probe: 250ug / mL streptavidin-coated superparamagnetic magnetic beads with a diameter of 1 μm, 50mM Tris, pH 7.0, 0.1% (V / V) NP-40, 1% (V / V) TritonX-100, 0.1% (V / V) SDS, 0.2uM biotinylated capture probe, wherein the capture probe is: HCV-DNA TCO: 5 '-AGACAGTAGTTCCTCACAGGGG-Biotin 3' (seq...
Embodiment 2
[0096] Embodiment 2, utilizing 2'-oxygen-methylated oligonucleotide (2'-OME-RNA) capture probe and 2'-deoxy oligonucleotide (DNA) capture probe to capture the comparison of HBV DNA target efficiency
[0097] Taking HBV DNA (sequence 1 in the sequence listing) as an example, using Biotin (biotin) as marker B and Strepavidin (streptavidin) as marker B specific binder A to detect 2'-oxo-methylated oligo The capture efficiency of the nucleic acid capture probe and the 2'-deoxy oligonucleotide capture probe to the DNA target nucleic acid, the specific method comprises the following steps:
[0098] 1. Reagent preparation
[0099] 1. the lysate (A) that has 2'-deoxyoligonucleic acid (DNA) capture probe: Same as embodiment 1, wherein, capture probe is: HBV-DNA TCO: 5'-CACGGGACCATGCAAGACCTGCACG-Biotin-3' (Sequence 6 in the sequence listing, 2'-deoxyoligonucleotide (DNA) capture probe).
[0100] 2. Lysate (B) with 2'-oxygen-methylated oligonucleotide (2'-OME-RNA) capture probe: same a...
Embodiment 3
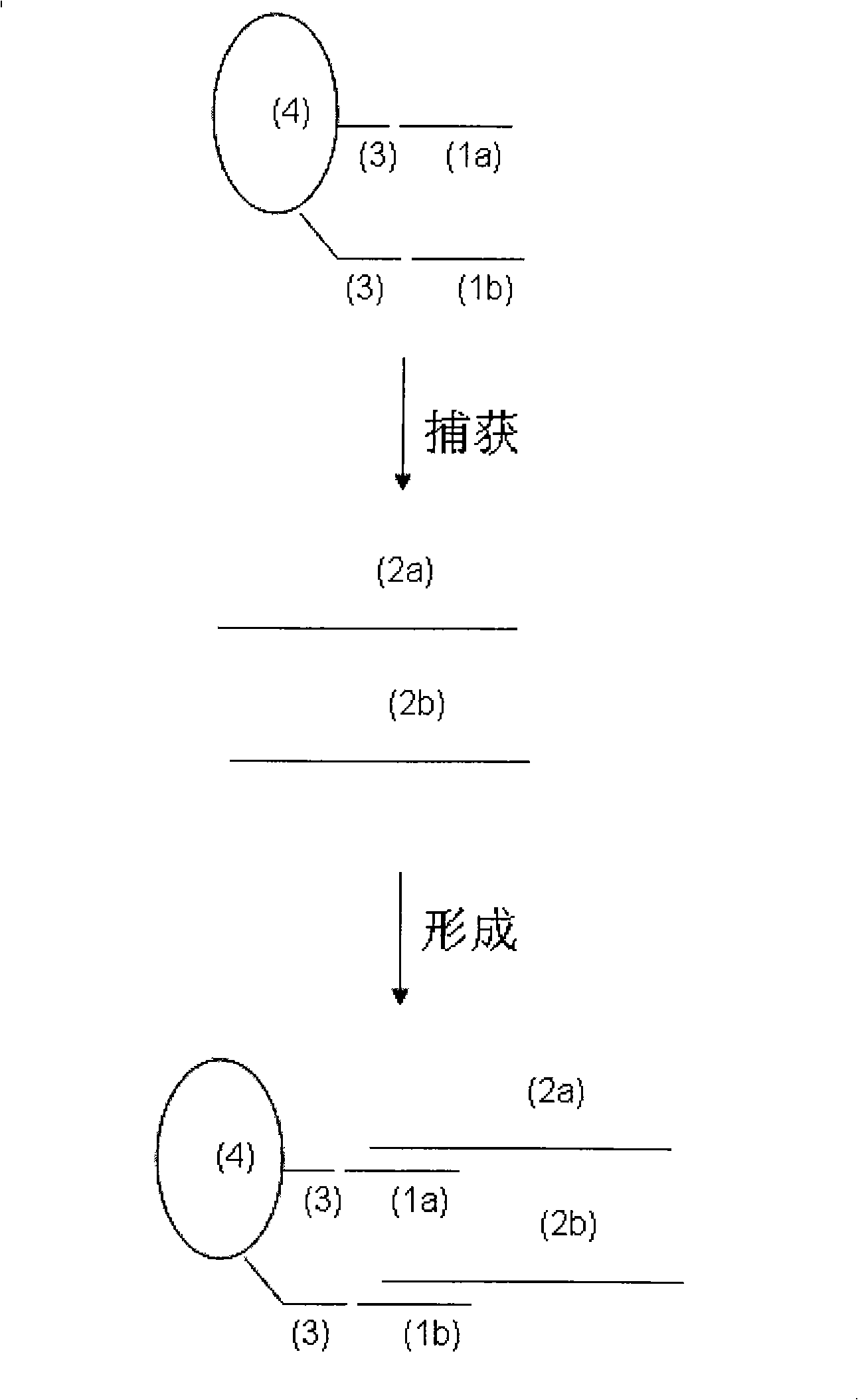
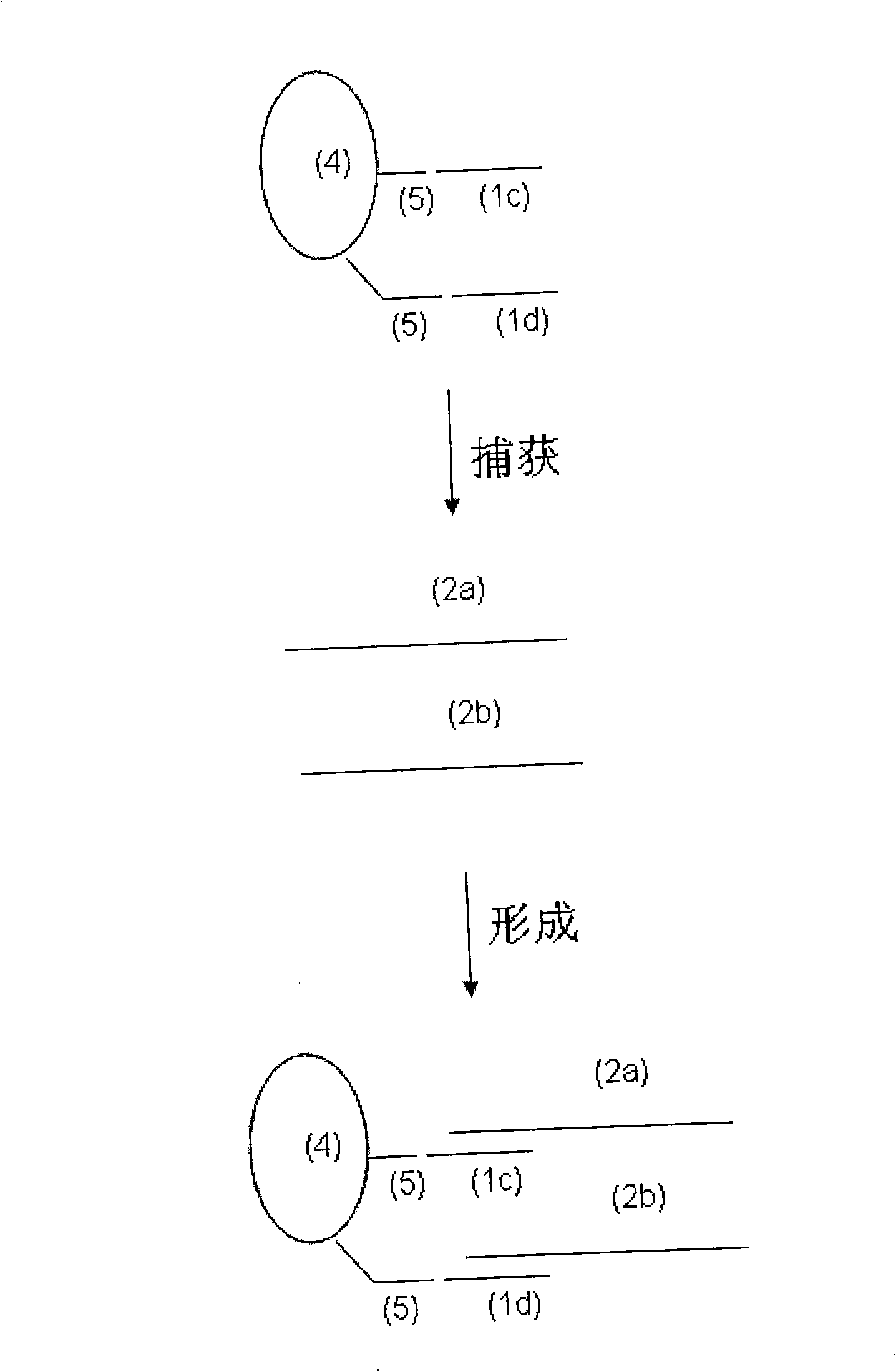
[0112] Embodiment 3, the 2'-deoxyoligonucleotide (DNA) capture probe that is used for HBV DNA capture in embodiment 2 and the 2'-oxygen-methylated oligonucleotide (2') that is used for HCV RNA capture in embodiment 1 '-OME-RNA) capture probes were used simultaneously, and the samples in Example 1 and Example 2 were detected in combination with SAT.
[0113] The detection method comprises the following steps:
[0114] 1. Reagent preparation
[0115] 1. the lysate with 2'-deoxy oligonucleotide (DNA) capture probe and 2'-oxygen-methylation oligonucleotide (2'-OME-RNA) capture probe: same as embodiment 1, wherein, The capture probes are: HBV-DNA-TCO: 5'-CACGGGACCATGCAAGACCTGCACG-Biotin3', HCV-RNA-TCO: 5'-AGACAGUAGUUCCUCAGGGG-Biotin-3'.
[0116] 2. Washing solution: 150mM NaCl solution.
[0117] 3. Preparation of HCV RNA samples and HBV DNA samples: S1 is HCV positive plasma containing about 100,000 copies / mL, S2 is ten-fold dilution of S1 with negative plasma, S3 is ten-fold di...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com