Vitreoscilla hemoglobin mutant and gene and application thereof
A technology of hemoglobin and hyaline bacteria, applied in the field of genetic engineering, can solve problems that are not necessarily optimal
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Construction of cloning and expression vectors
[0035] Utilize the Vector NTI10 software of Invitrogen Company, design primer VH1 and VH2 according to the vgb of 1.4kb and its promoter sequence reported in literature Dikshit KL, WebsterDA.Gene, 1988,70:377-386:
[0036] VH1:5'-GC TCTAGA TTATTCAACCGCT
[0037] wxya
[0038] VH2: 5'-CG GAATTC TATGTTAGACCAGCAAAACCA-3' (SEQ ID No. 4)
[0039] EcoR I
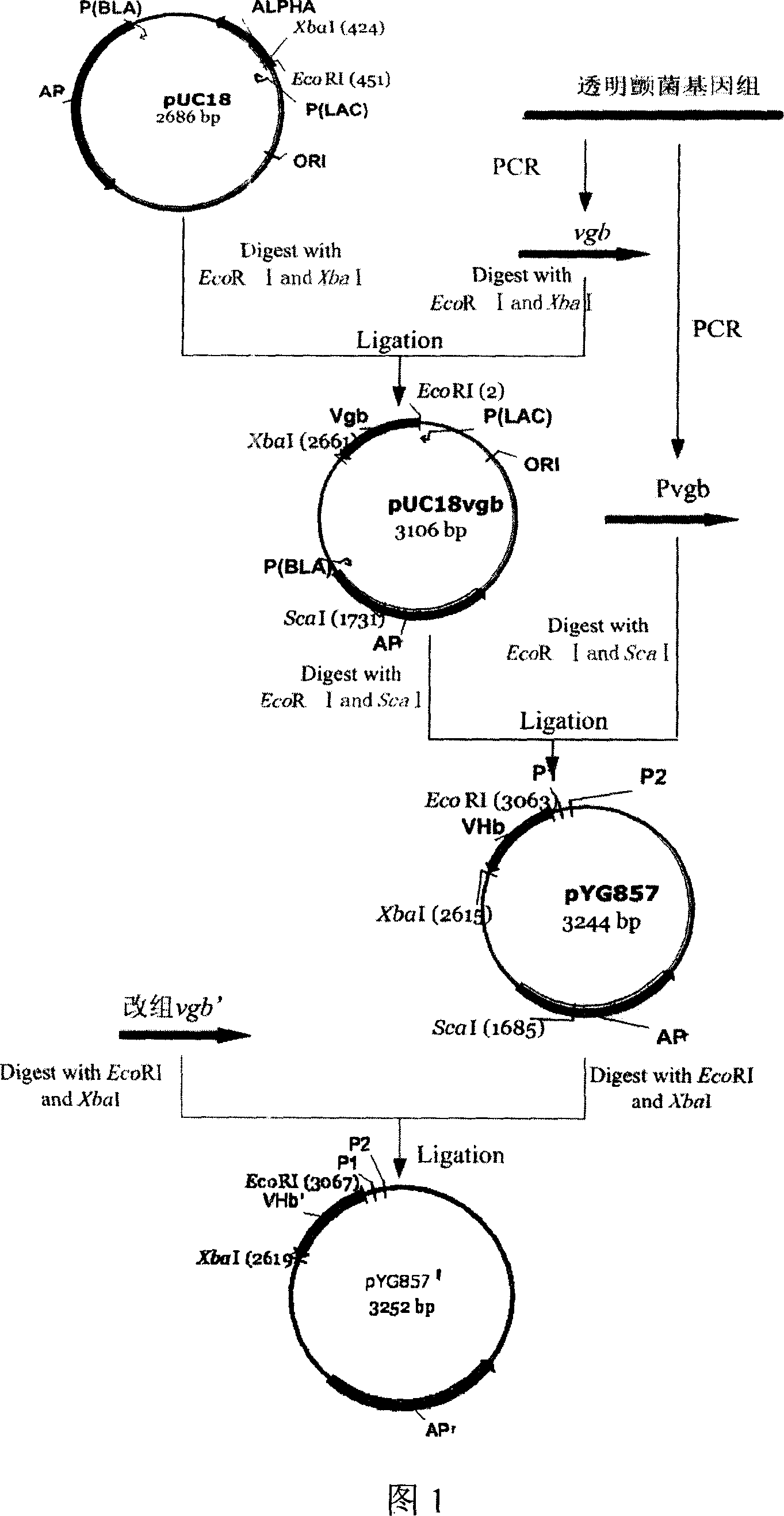
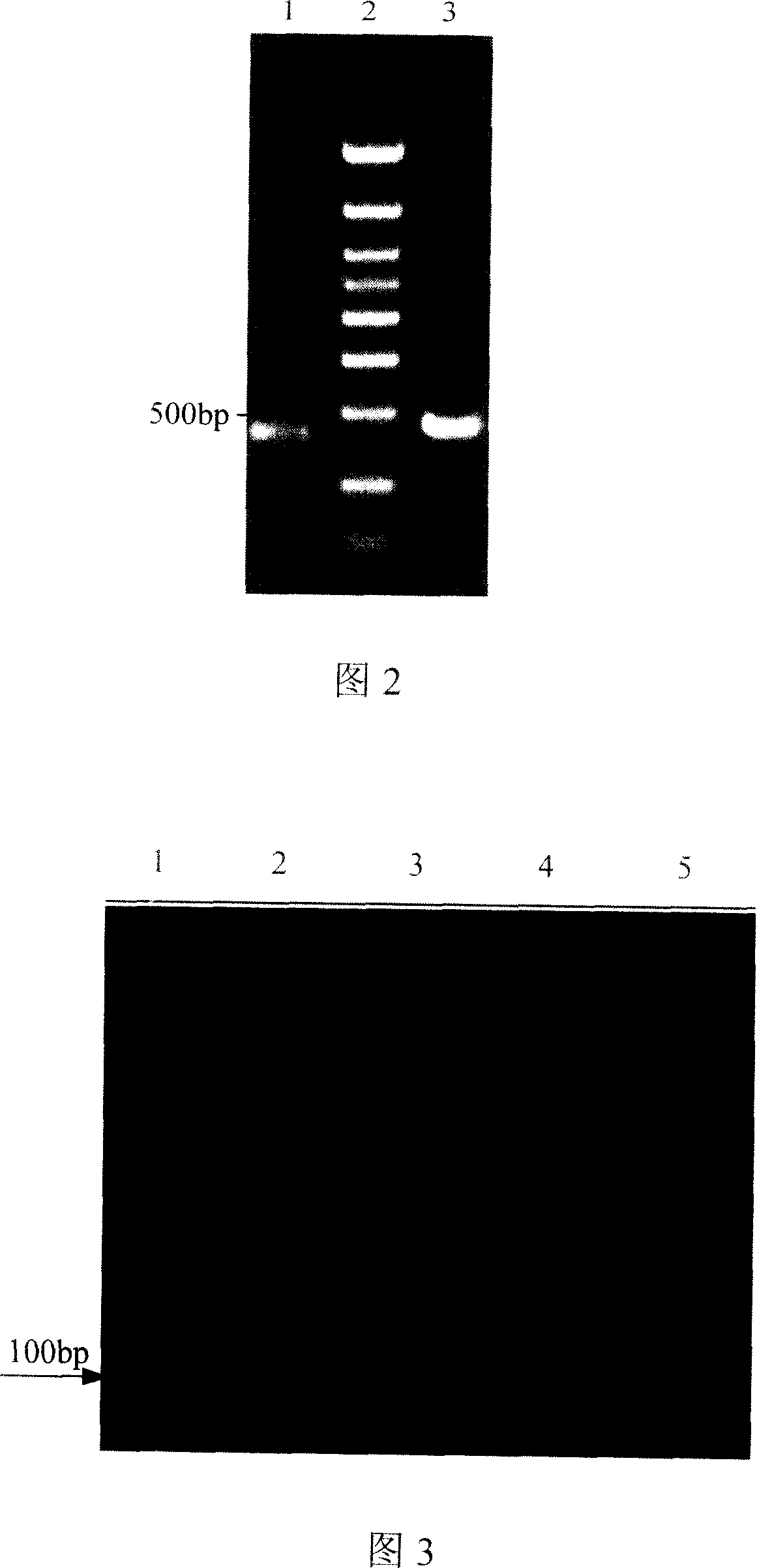
[0040] Using the genome of Vitreoscilla filiformis ATCC43191 as a template, VH1 and VH2 were used as primers to amplify the vgb structural gene. The reaction conditions were: 94°C for 5 minutes, 94°C for 30s, 57°C for 30s, and 72°C for 40s. After 30 cycles, Extend at 72°C for 5 min. The PCR product was separated by 1% agarose gel electrophoresis, and a fragment around 500 bp was recovered. After being digested by EcoR I and XbaI, it was connected into pUC18 which had been digested by the same double enzyme to obtain pUC18vgb.
[0041] Also design primers Pvg...
Embodiment 2
[0050] Embodiment 2 utilizes error-prone PCR to obtain vgb gene shuffling material
[0051] Use pYG857 template, VH1 and VH2 as primers to amplify vgb, add MgCl to the PCR system 2 and MnCl 2 , so that Mg 2+ and Mn 2+ The final concentrations were 7.5mmol / L and 0.5mmol / L respectively, and the PCR reaction conditions and product processing were as described in Example 1. The error-prone PCR product was recovered by electrophoresis (Fig. 2), and double-digested with EcoR I and Xba[, and the digested product was connected to pYG857 through the same digestion to obtain the mutant gene vector pYG857EP, which was transformed into E.coli DH5α competent cells, Spread on LB agar plate containing ampicillin and culture overnight at 37°C.
[0052] Randomly pick a single transformant colony from the transformation plate, insert it into a 15ml test tube containing 5ml LB liquid medium, add antibiotics, seal the test tube mouth with a rubber cap, and cultivate at 37°C, 230r / min for abou...
Embodiment 3
[0053] Example 3 DNA shuffling of vgb gene
[0054] (1) DNA fragmentation reaction
[0055] Using pYG857 and 4 pYG857EP screened by error-prone PCR as templates, use the conditions described in Example 1 to amplify the original gene vgb and mutant gene vgb', recover the target fragment by electrophoresis, and dissolve it in an appropriate volume of ddH 2 O middle. In a 20 μl system, add about 0.5 μg of each of the five recovered products and 0.5 UDNase I, react at 25°C for 14-20min, and inactivate the enzyme at 90°C for 10min. The degree of fragmentation of the product was detected by agarose gel electrophoresis. It can be seen from Fig. 3 that when the digestion reaction is carried out for about 18 minutes, ideal fragments of 50 bp can be obtained. Cut out the gel block where the fragments are concentrated near 50bp, and recover it with the small fragment DNA rapid recovery kit.
[0056] (2) PCR without primers
[0057] Take 40 μl of small fragment recovery products dire...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com