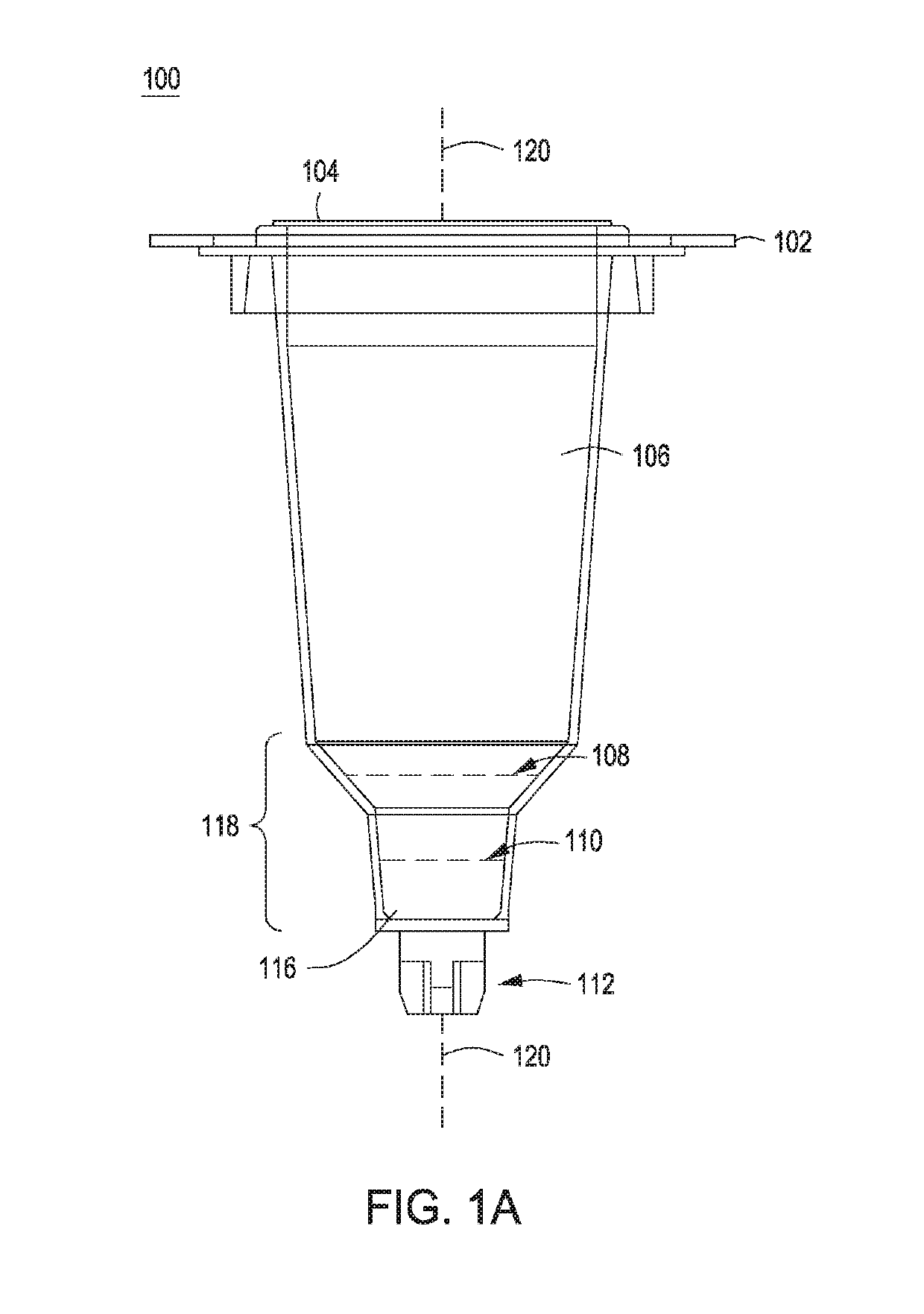
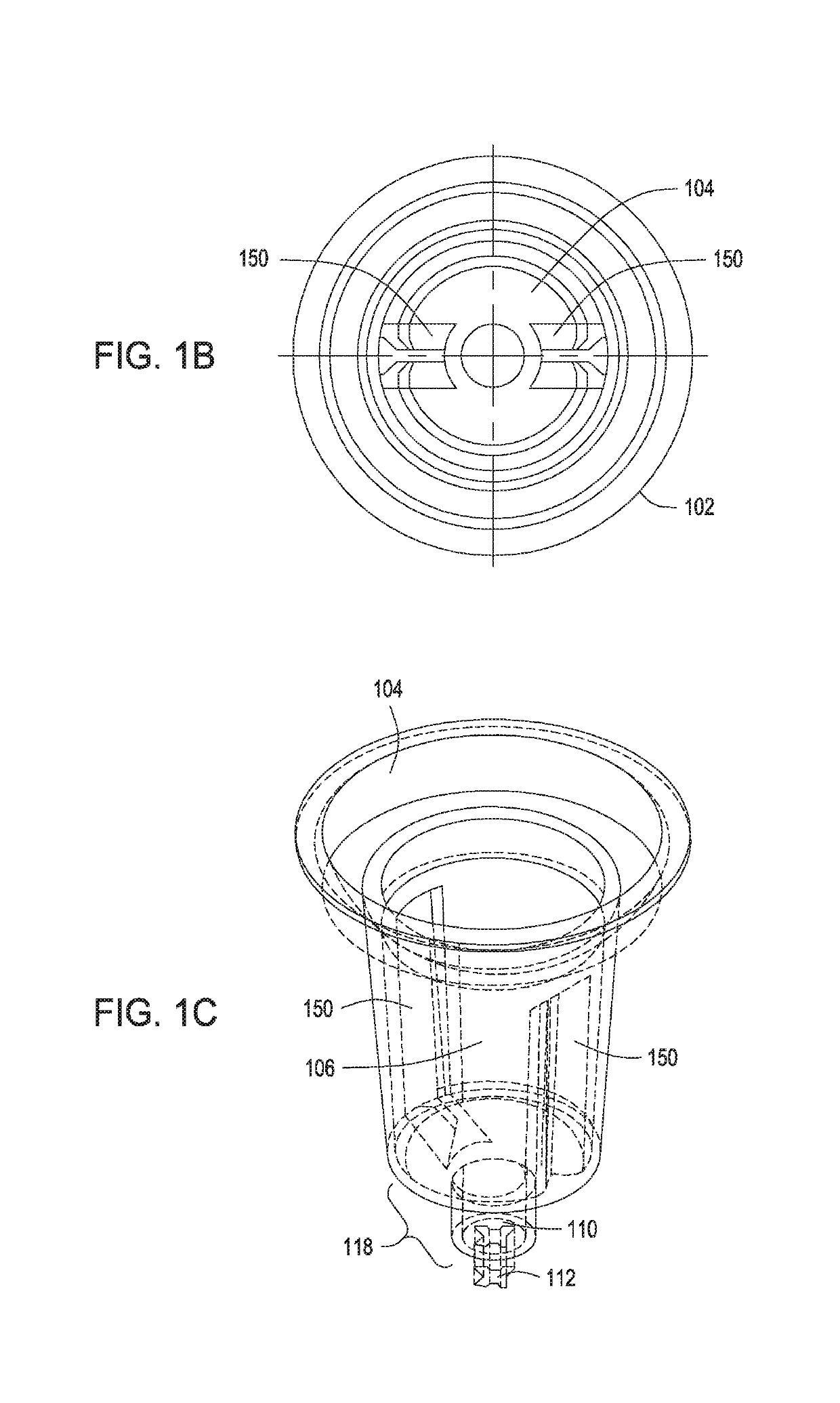
Methods for controlling the growth of prokaryotic and eukaryotic cells
a technology for prokaryotic cells and eukaryotic cells, applied in biochemistry apparatus and processes, specific use bioreactors/fermenters, after-treatment of biomass, etc., can solve the problems of contaminated cell culture, time-consuming and labor-intensive procedures, and human intervention
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example i
Growth in the Cell Growth Module
[0113]One embodiment of the cell growth device as described herein was tested against a conventional cell shaker shaking a 5 ml tube and an orbital shaker shaking a 125 ml baffled flask to evaluate cell growth in bacterial and yeast cells. Additionally, growth of a bacterial cell culture and a yeast cell culture was monitored in real time using an embodiment of the cell growth device described herein.
[0114]In a first example, 20 ml EC23 cells (E. coli cells) in LB were grown in a 35 ml rotating growth vial with a 2-paddle configuration at 30° C. using the cell growth device as described herein. The rotating growth vial was spun at 600 rpm and oscillated (i.e., the rotation direction was changed) every 1 second. In parallel, 5 ml EC23 cells in LB were grown in a 5 ml tube at 30° C. and were shaken at 750 rpm. OD600 was measured at intervals using a NanoDrop™ spectrophotometer (Thermo Fisher Scientific). The results are shown in FIG. 8. The rotating gro...
example ii
Fully-Automated Singleplex RGN-Directed Editing Run
[0118]Singleplex automated genomic editing using MAD7 nuclease was successfully performed with an automated multi-module instrument of the disclosure. See U.S. Pat. No. 9,982,279.
[0119]An ampR plasmid backbone and a lacZ_F172* editing cassette were assembled via Gibson Assembly® into an “editing vector” in an isothermal nucleic acid assembly module included in the automated instrument. lacZ_F172 functionally knocks out the lacZ gene. “lacZ_F172*” indicates that the edit happens at the 172nd residue in the lacZ amino acid sequence. Following assembly, the product was de-salted in the isothermal nucleic acid assembly module using AMPure beads, washed with 80% ethanol, and eluted in buffer. The assembled editing vector and recombineering-ready, electrocompetent E. Coli cells were transferred into a transformation module for electroporation. The cells and nucleic acids were combined and allowed to mix for 1 minute, and electroporation w...
example iii
Fully-Automated Recursive Editing Run
[0122]Recursive editing was successfully achieved using the automated multi-module cell processing system. An ampR plasmid backbone and a lacZ_V10* editing cassette were assembled via Gibson Assembly® into an “editing vector” in an isothermal nucleic acid assembly module included in the automated system. Similar to the lacZ_F172 edit, the lacZ_V10 edit functionally knocks out the lacZ gene. “lacZ_V10” indicates that the edit happens at amino acid position 10 in the lacZ amino acid sequence. Following assembly, the product was de-salted in the isothermal nucleic acid assembly module using AMPure beads, washed with 80% ethanol, and eluted in buffer. The first assembled editing vector and the recombineering-ready electrocompetent E. Coli cells were transferred into a transformation module for electroporation. The cells and nucleic acids were combined and allowed to mix for 1 minute, and electroporation was performed for 30 seconds. The parameters fo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com