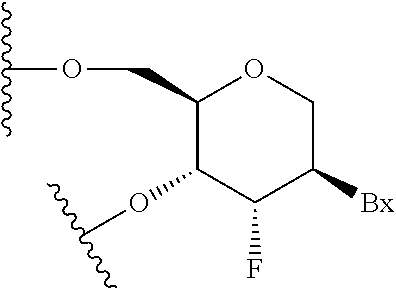
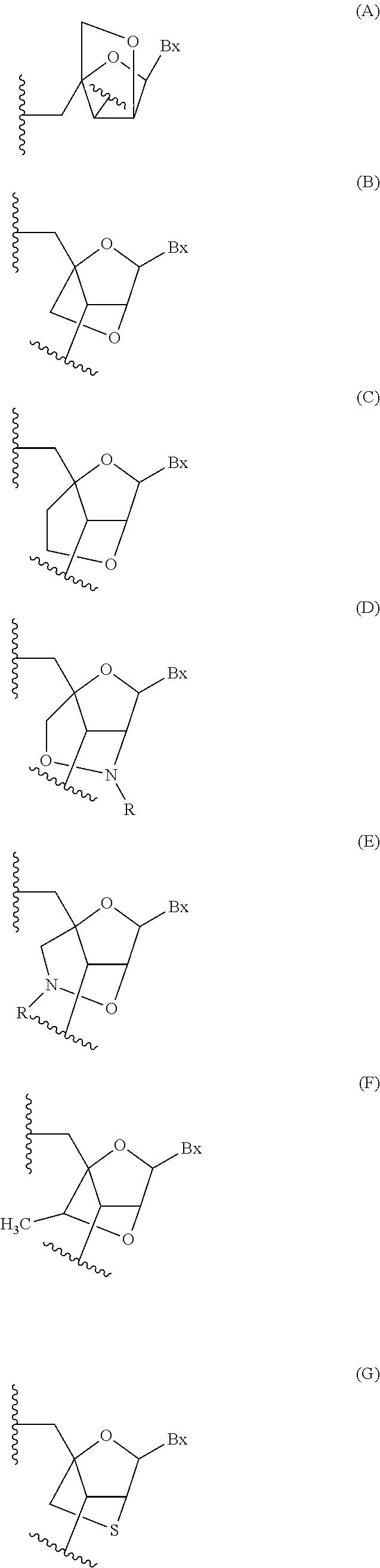
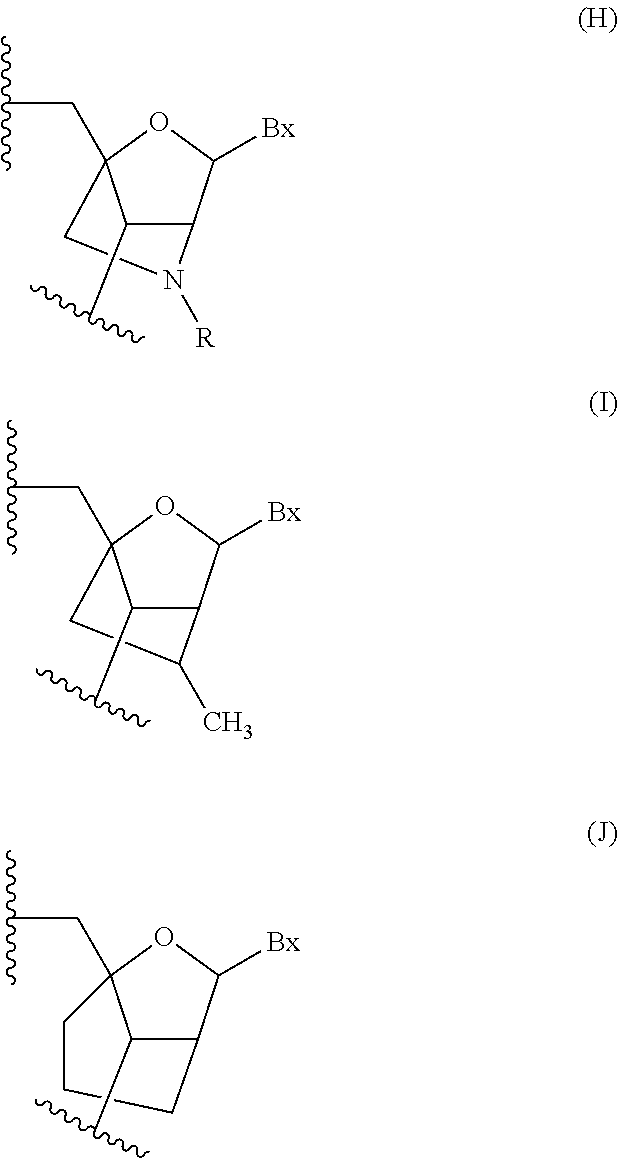
Modulation of alpha synuclein expression
a technology of alpha synuclein and modulation, which is applied in the direction of muscular disorders, organic chemistry, drug compositions, etc., can solve the problems of lack of acceptable options for treating such neurodegenerative disorders
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Antisense Inhibition of Human Alpha-Synuclein (SNCA) in HuVEC Cells
[0287]Antisense oligonucleotides targeted to an SNCA nucleic acid were tested for their effects on SNCA mRNA in vitro. Cultured HuVEC cells at a density of 5,000 cells per well were transfected using LipofectAMINE2000® reagent with 10 nM antisense oligonucleotide. After a treatment period of approximately 24 hours, RNA was isolated from the cells and SNCA mRNA levels were measured by quantitative real time PCR using the human primer probe set RTS2621 (forward sequence ACGAACCTGAAGCCTAAGAAATATCT, designated herein as SEQ ID NO: 8; reverse sequence GAGCACTTGTACAGGATGGAACAT, designated herein as SEQ ID NO: 9, probe sequence TGCTCCCAGTTTCTTGAGATCTGCTGACA, designated herein as SEQ ID NO: 10). SNCA mRNA levels were adjusted according to total RNA content, as measured by RIBOGREEN. Results are presented as percent inhibition of SNCA, relative to untreated control cells.
[0288]The chimeric antisense oligonucleotides in Tables...
example 2
Dose-Dependent Antisense Inhibition of Human SNCA in HuVEC Cells
[0294]Eleven gapmers, exhibiting over 84 percent or greater in vitro inhibition of human SNCA in the study described in Example 1, were tested at various doses in HuVEC cells. Cells were plated at a density of 6,000 cells per well and transfected using LipofectAMINE2000® reagent with 0.08 nM, 0.25 nM, 0.74 nM, 2.22 nM, 6.67 nM, and 20.00 nM concentrations of antisense oligonucleotide, as specified in Table 4. After a treatment period of approximately 16 hours, RNA was isolated from the cells and SNCA mRNA levels were measured by quantitative real-time PCR. Human SNCA primer probe set RTS2621 (described herein above in Example 1) was used to measure mRNA levels. SNCA mRNA levels were adjusted according to total RNA content, as measured by RIBOGREEN. Results are presented as percent inhibition of SNCA, relative to untreated control cells. As illustrated in Table 4, SNCA mRNA levels were reduced in a dose-dependent manner ...
example 3
Dose-Dependent Antisense Inhibition of Human SNCA in SH-SY5Y Cells
[0295]Gapmers were selected from the study described in Example 2 and tested at various doses in SH-SY5Y cells. Cells were plated at a density of 20,000 cells per well and transfected using electroporation with 5 μM, 10 μM, and 20 μM concentrations of antisense oligonucleotide, as specified in Table 5. After a treatment period of approximately 16 hours, RNA was isolated from the cells and SNCA mRNA levels were measured by quantitative real-time PCR. Human SNCA primer probe set RTS2620 (forward sequence GGTGCTTCCCTTTCACTGAAGT, designated herein as SEQ ID NO: 89; reverse sequence ACATCGTAGATTGAAGCCACAAAA, designated herein as SEQ ID NO: 90, probe sequence AATACATGGTAGCAGGGTCTTTGTGTGCTGTG, designated herein as SEQ ID NO: 91) was used to measure mRNA levels. SNCA mRNA levels were adjusted according to total RNA content, as measured by RIBOGREEN. Results are presented as percent inhibition of SNCA, relative to untreated co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com