Hydrolysis Probes
a technology of hydrolysis probes and probes, applied in the field of molecular biology, can solve the problems of difficult to quantify the starting template, real-time pcr technology, unparallel amplification and precision capability, etc., and achieve the effect of determining the amplification efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
A. Hydrolysis Probes
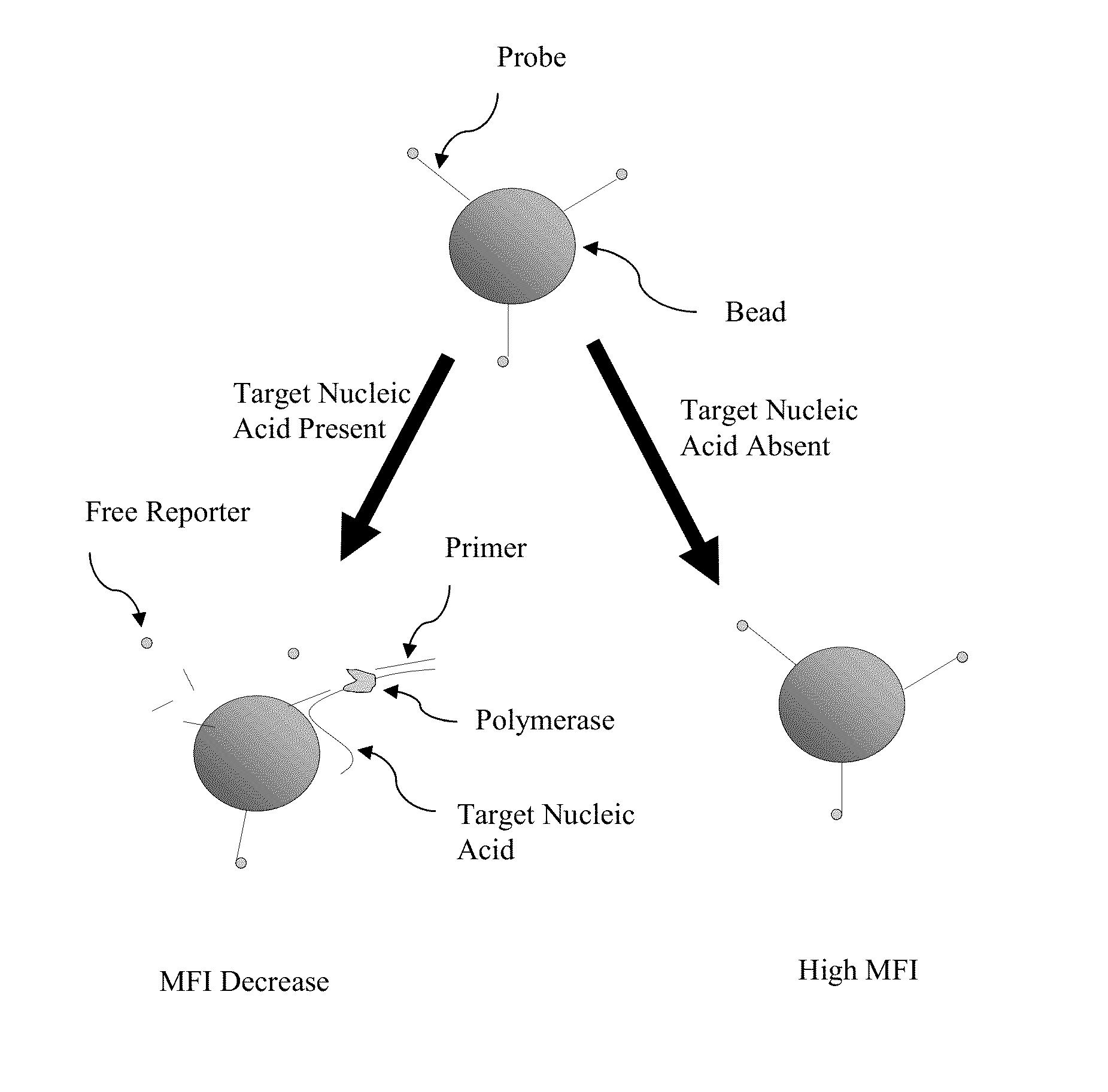
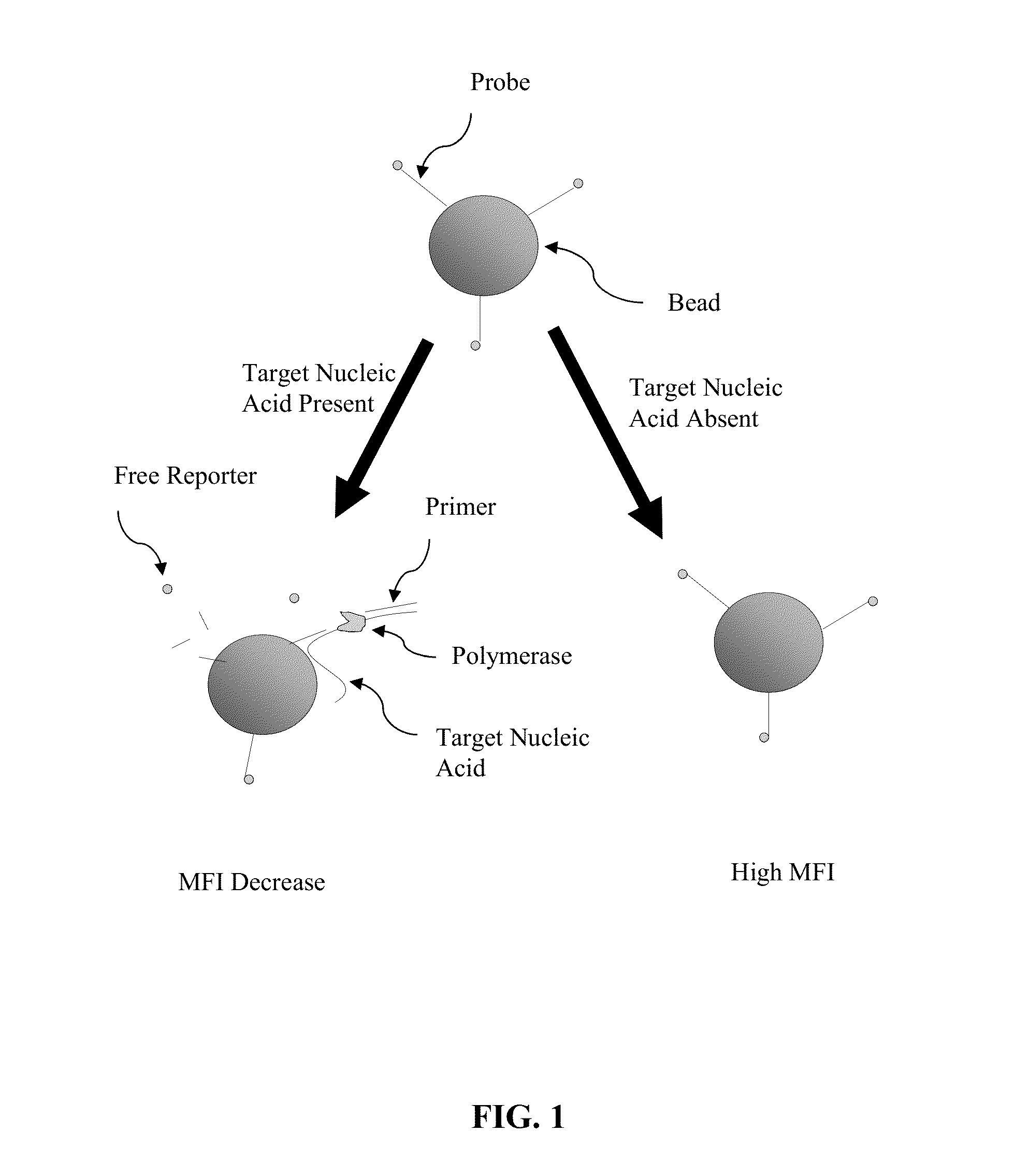
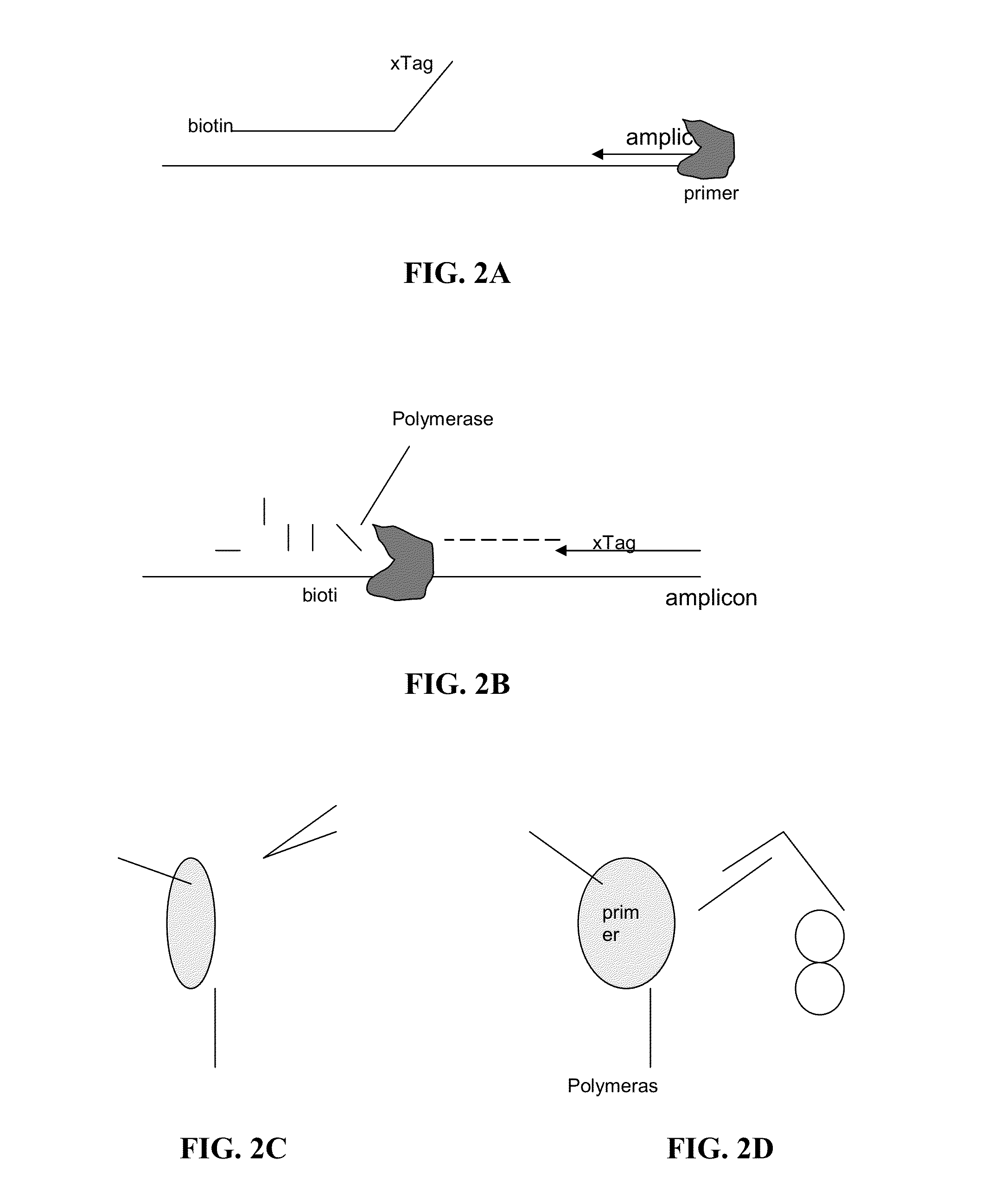
[0053]Certain aspects of the present invention employ hydrolysis probes for the detection of nucleic acids. Hydrolysis probes take advantage of the 5′ exonuclease activity of some polymerases. During the extension or elongation phase of a PCR reaction, a polymerase, such as Taq polymerase, uses an upstream primer as a binding site and then extends. The hydrolysis probe is then cleaved during polymerase extension at its 5′ end by the 5′-exonuclease activity of the polymerase.
[0054]However, the process of cleaving the 5′ end of the probe need not require amplification or extension of the target sequence (see, e.g., U.S. Pat. No. 5,487,972, incorporated herein by reference). This is accomplished by placing the probe in close proximity to the upstream primer on the target sequence such that binding of the nucleic acid polymerase to the 3′ end of the primer automatically puts the polymerase in contact with the 5′ end of the probe. Because polymerization is not require...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Tm | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com