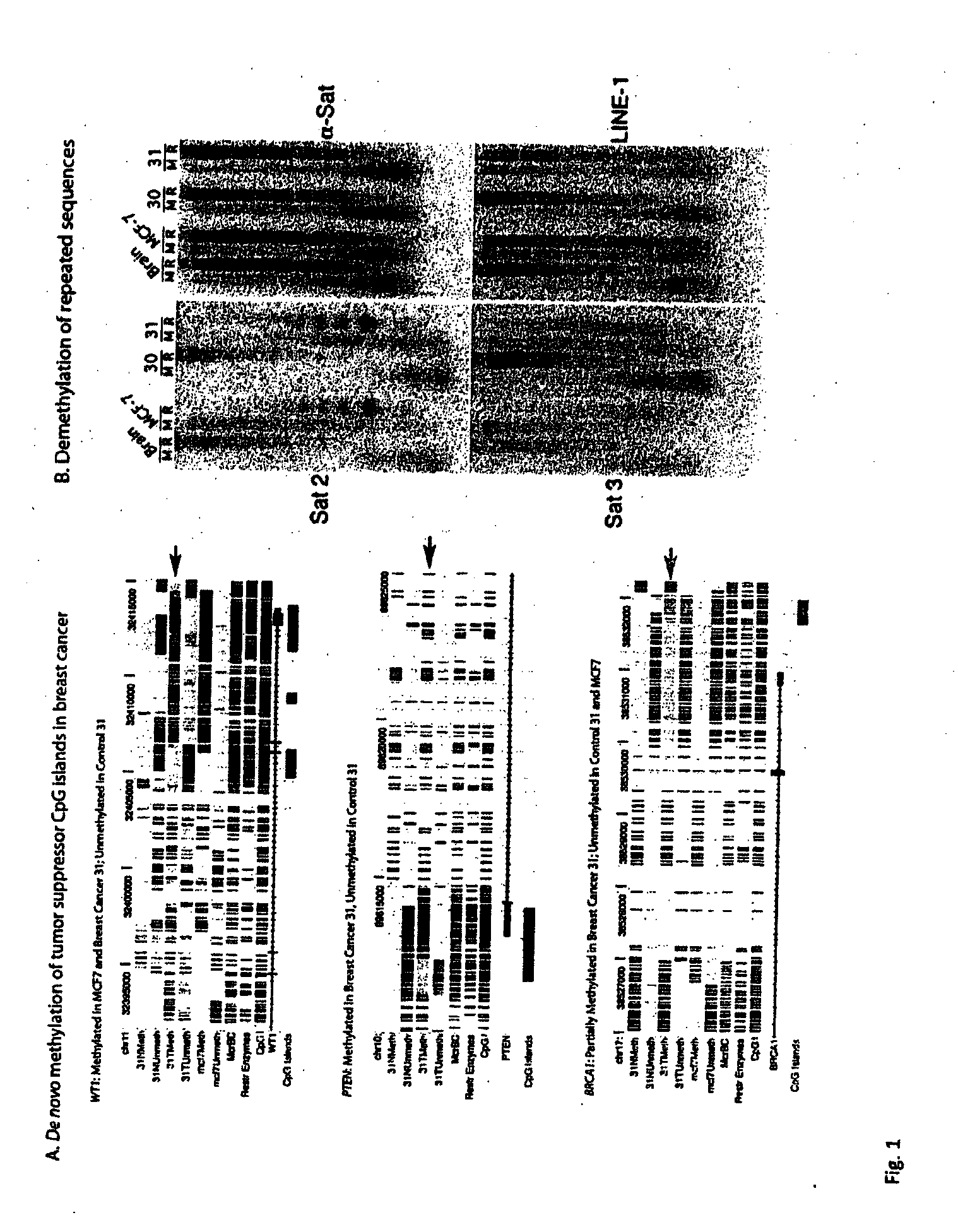
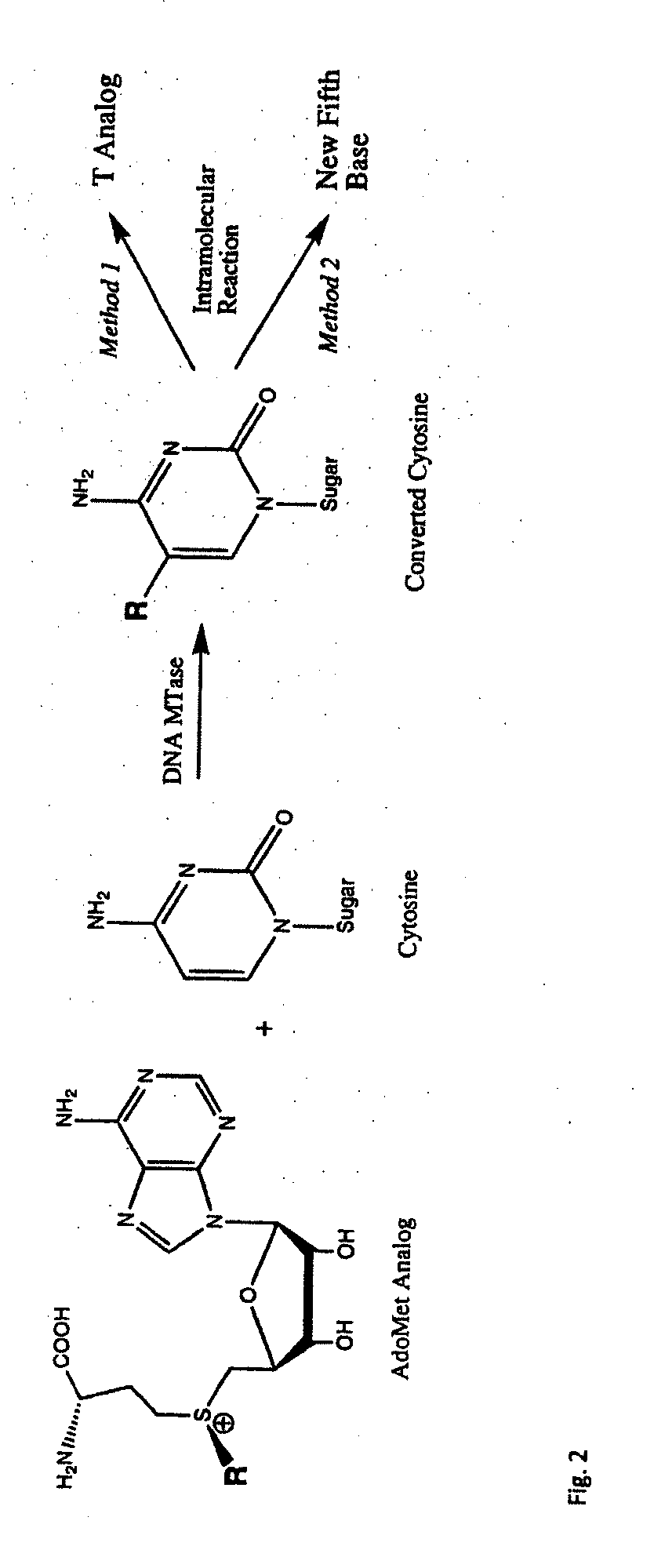
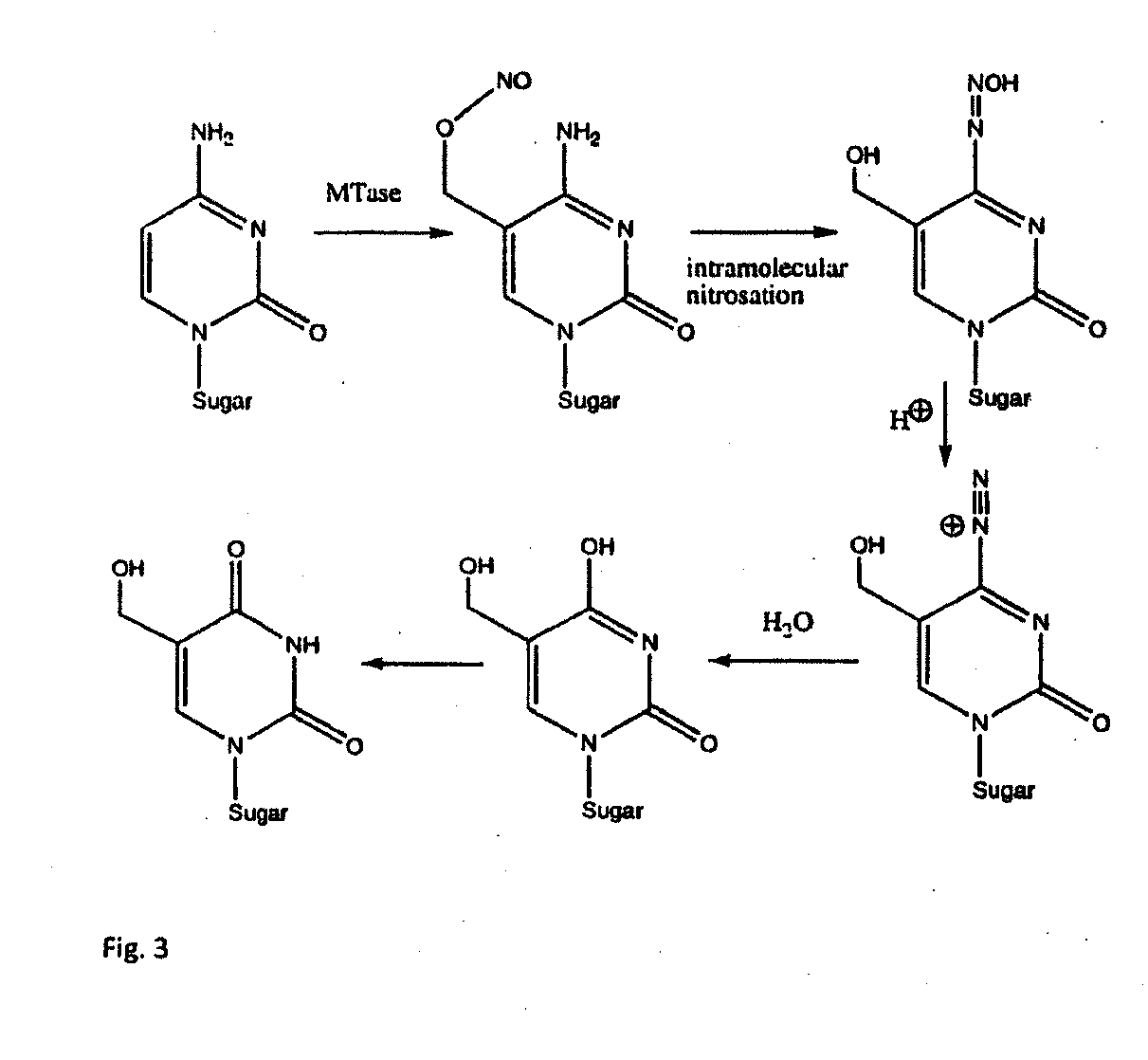
Universal methylation profiling methods
a technology of methylation profiling and profiling methods, applied in the field of universal methylation profiling methods, can solve the problems of difficult to discern abnormality of genomic methylation patterns found in human diseases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
Terms
[0056]As used herein, and unless stated otherwise, each of the following terms shall have the definition set forth below.
A—Adenine;
C—Cytosine;
[0057]DNA—Deoxyribonucleic acid;
G—Guanine;
[0058]RNA—Ribonucleic acid;
T—Thymine; and
U—Uracil.
[0059]“Nucleic acid” shall mean any nucleic acid molecule, including, without limitation, DNA, RNA and hybrids thereof. The nucleic acid bases that form nucleic acid molecules can be the bases A, C, G, T and U, as well as derivatives thereof. Derivatives of these bases are well known in the art, and are exemplified in PCR Systems, Reagents and Consumables (Perkin Elmer Catalogue 1996-1997, Roche Molecular Systems, Inc., Branchburg, N.J., USA).
[0060]“Type” of nucleotide refers to A, G, C, T or U. “Type” of base refers to adenine, guanine, cytosine, uracil or thymine.
[0061]“Mass tag” shall mean a molecular entity of a predetermined size which is capable of being attached by a cleavable bond to another entity.
[0062]“Solid substrates” shall mean any su...
PUM
| Property | Measurement | Unit |
|---|---|---|
| structure | aaaaa | aaaaa |
| genome stability | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com