Methods for identifying and isolating unique nucleic acid sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Characterization of Minute Differences Between Genomes of Strains of Penicillium nalgiovense Using Subtractive Suppression Hybridization (SSH) Without Cloning
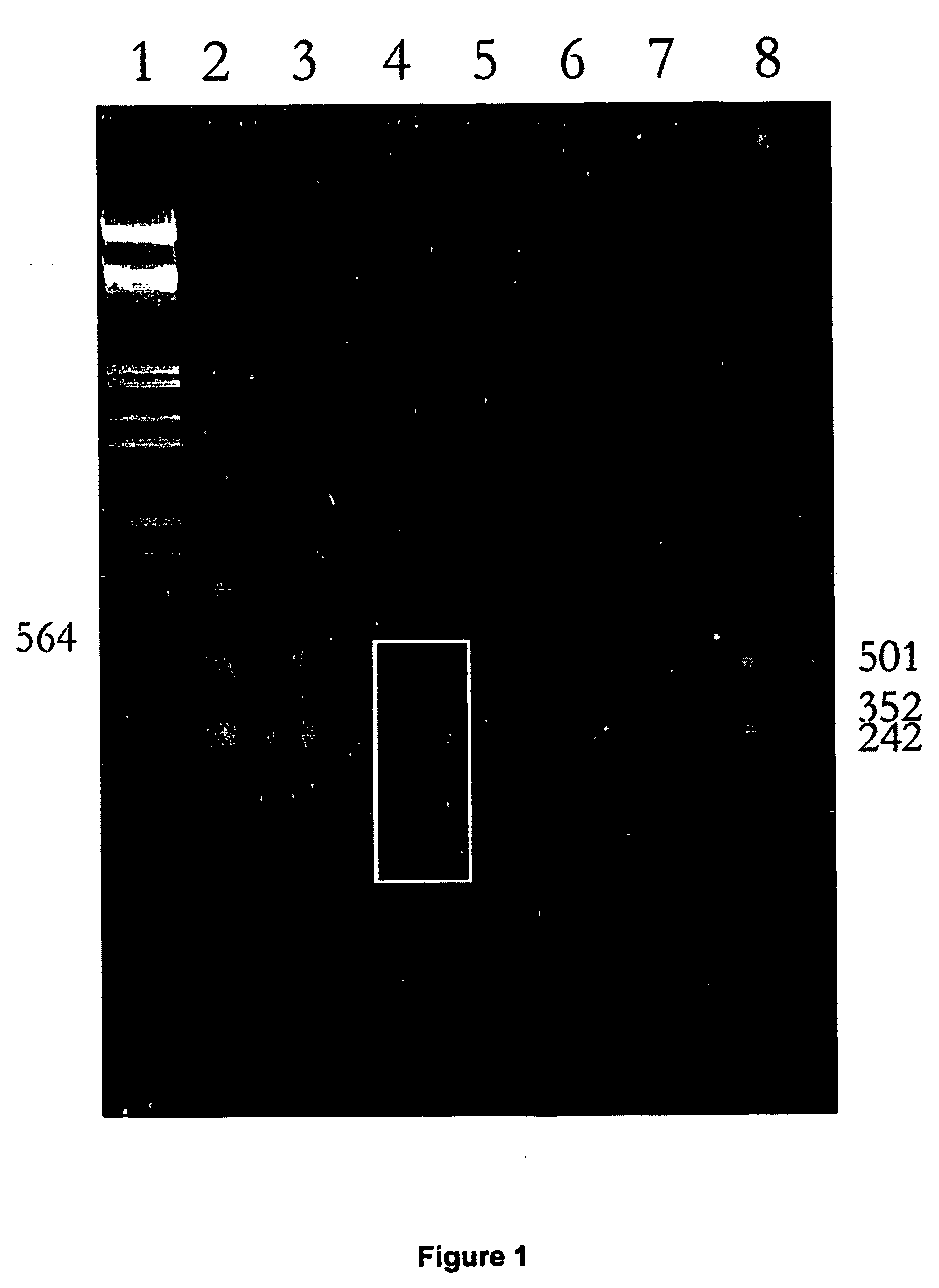
[0075]Penicillium nalviogensis (designated as BFE) was supplied by Bundesforschungsanstalt für Emährung, Karlsruhe; BFE. Strains BFE 19 and BFE 20 were used as tester organisms and were genetically modified by cloning the vector p3SR2 and pKW 100 (without further accessible data) which were incorporated into the genome at different locations. BFE 66 as well as BFE 328 were used as driver organisms. The molds were cultivated in malt medium and malt agar petridishes at 25° C. in the dark. Bacterial contamination was inhibited by adding Kanamycin (50 μg / ml) to the medium. DNA isolation was carried out according to Waver et al. (1995).
Subtractive Suppression Hybridization
[0076] The SSH procedure was performed with slight modifications as described (Diatchenko et al. (1996) using RSAI restriction endonuclease (Amersham Life Scie...
example 2
SSH (Subtractive Suppression Hybridization) Using Human Genomic DNA as Template
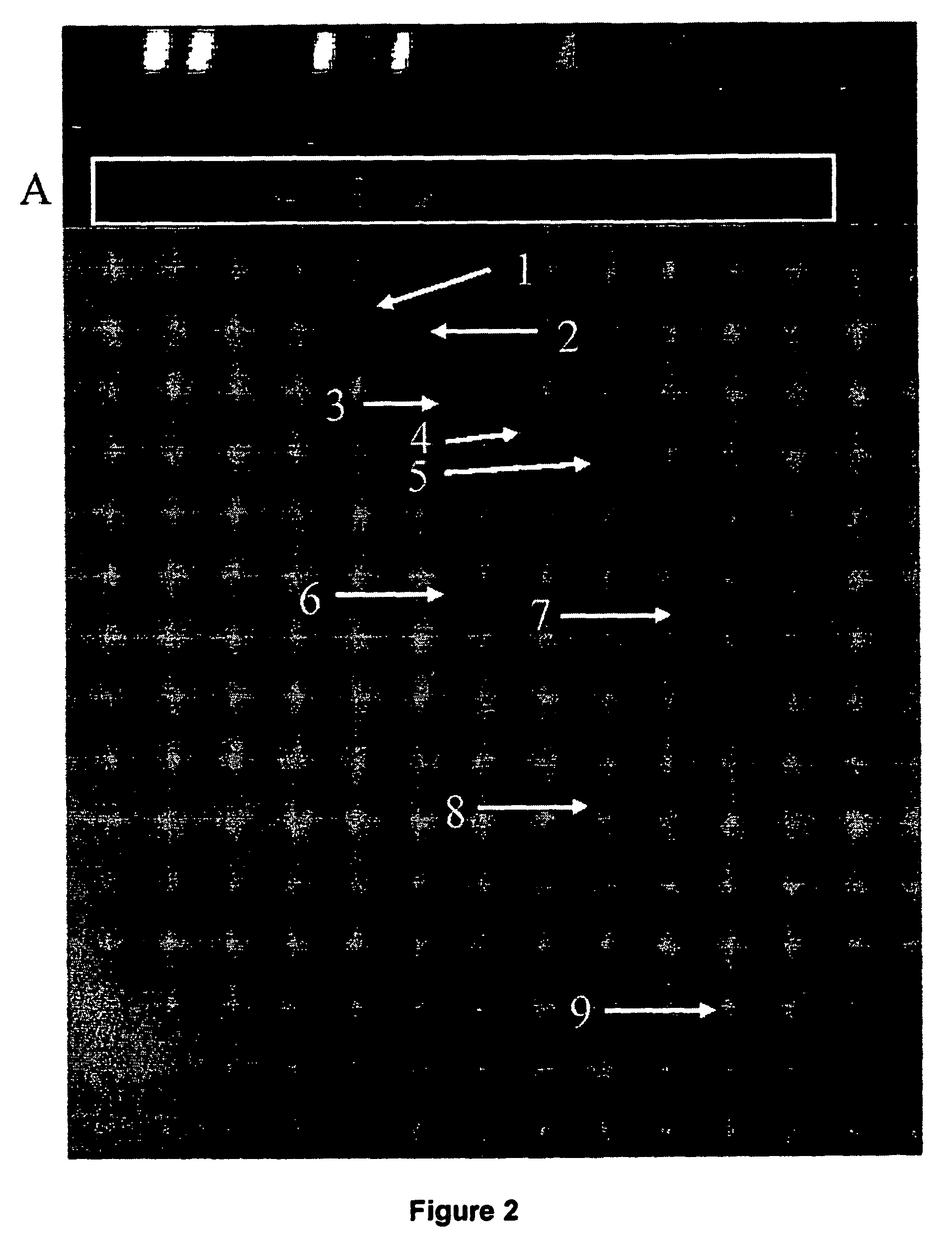
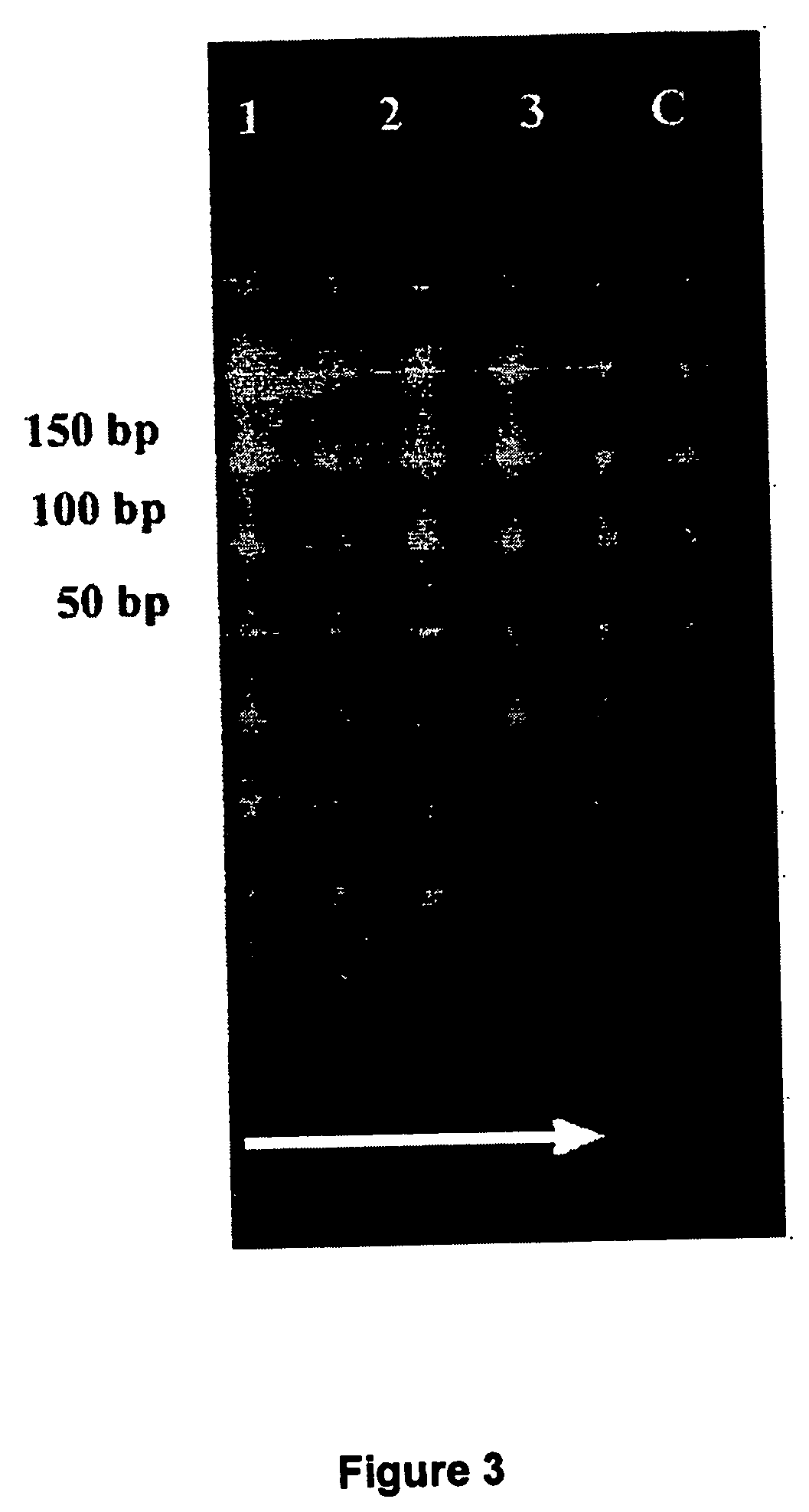
[0082] DNA isolation, restriction enzyme digestion of the DNA as well as the subtractive suppression hybridization was carried out as described in Example 1. First hybridization and second hybridization was performed with slightly modifications. The first subtractive hybridization was carried out with 1.5 μl tester 1 or tester 2R and 1 μl hybridization buffer plus 1.5 μl driver DNA (excess 30 fold) (Clontech). The samples were covered with mineral oil and hybridized at 63° C. for 16 h with addition of fresh denatured driver DNA after 2 and 5 h due to the high complexity of the human genome. Second hybridization was performed as described in Example 1.
[0083] Polymerase chain reaction: enrichment of tester specific sequences was carried out as described in Example 1 with slightly modifications. The template DNA was increased prior to suppression PCR attempt by diluting DNA samples 1:10 and applying 10, 5,...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Length | aaaaa | aaaaa |
| Electrical resistance | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com