Ehrlichia chaffeensis 28 kDa outer membrane protein multigene family
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 2
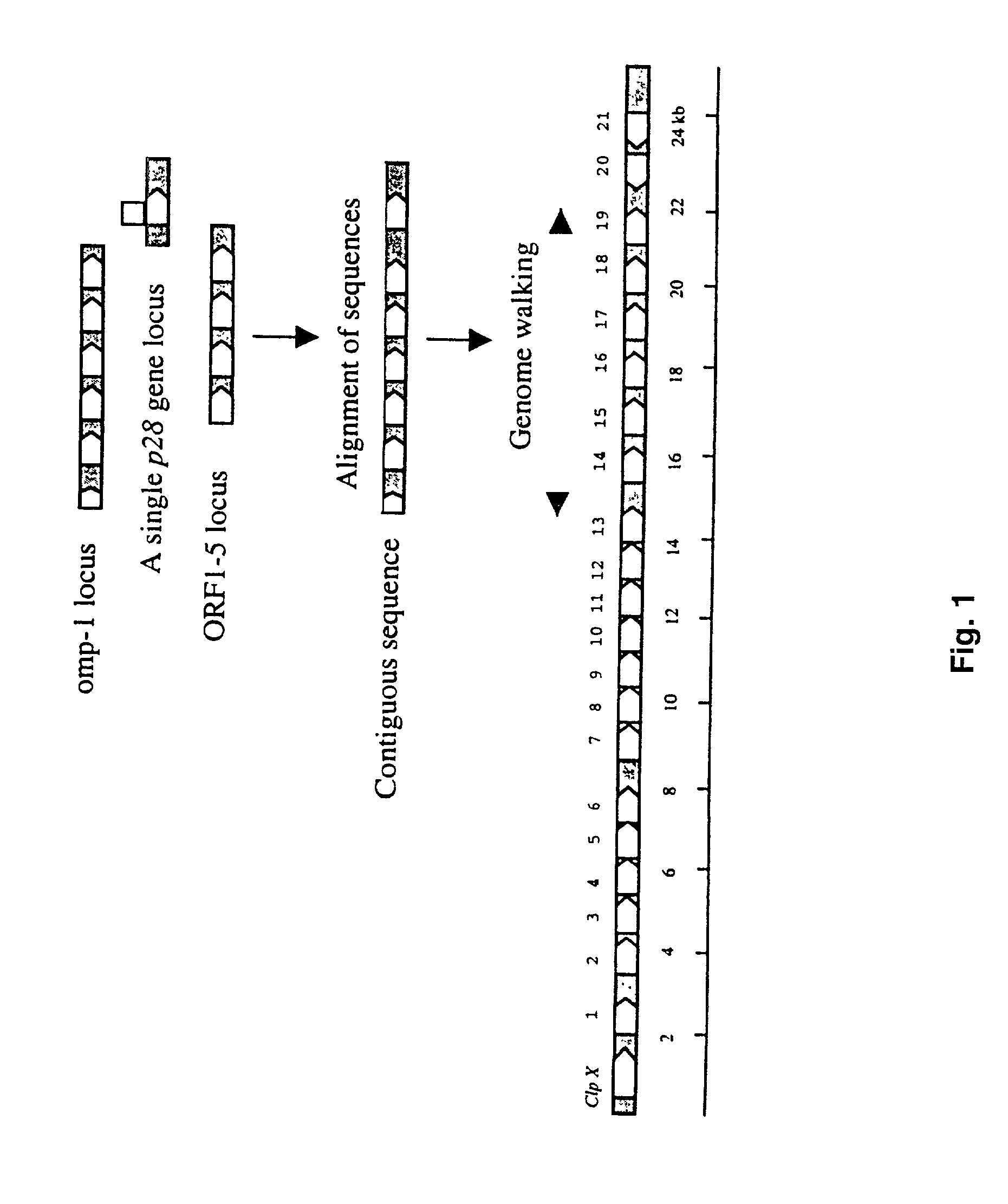
[0060] PCR Amplification of the p28 Multigene Locus
[0061] Ehrlichia chaffeensis genomic DNA was prepared by using an IsoQuick Nucleic Acid Extraction Kit (ORCA Research Inc., Bothell, Wash.) according to the instructions of the manufacturer. The unknown sequences of the p28 multigene locus were amplified by PCR using the Universal GenomeWalker Kit (Clontech Laboratories, Inc., Palo Alto, Calif.). Briefly, the E. chaffeensis genomic DNA was digested respectively with Dra I, EcoR V, Pvu II, Sca I, and Stu I. The enzymes were chosen because they generated blunt ended DNA fragments to ligate with the blunt-end of the adapter. The digested E. chaffeensis genomic DNA fragments were ligated with a GenomeWalker Adapter, which had one blunt end and one end with 5' overhang. The ligation mixture of the adapter and E. chaffeensis genomic DNA fragments was used as template for PCR. Initially, the p28 gene-specific primer amplified the known DNA sequence and extended into the unknown adjacent ge...
example 4
[0064] Gene Analysis
[0065] DNA sequences and deduced amino acid sequences were analyzed using DNASTAR software (DNASTAR, Inc., Madison, Wis.). The signal sequence of the deduced protein was analyzed by using the PSORT program, which predicts the presence of signal sequences (McGeoch, 1985, Von Heijne, 1986) and detects potential transmembrane domains (Klein, 1985). Phylogenetic analysis was performed by the maximum parsimony method of the PAUP 4.0 software (Sunderland Mass.: Sinauer Associates, 1998). Bootstrap values for the consensus tree were based on analysis of 1000 replicates.
example 5
[0066] DNA Sequence Accession Numbers
[0067] The DNA sequences of the E. chaffeensis p28 genes were assigned GenBank accession numbers: AF230642 for the DNA locus of the p28-1 to p28-13 and AF230643 for the DNA locus of p28-20 and p28-21.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Angle | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com