Method for auxiliary screening of high-oil corn and dedicated quantitative character gene locus therefor
A technology for high-oil corn and corn, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve the problem of difficulty in tracking the genetic effect of a single QTL, and achieve the effect of a quick selection method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
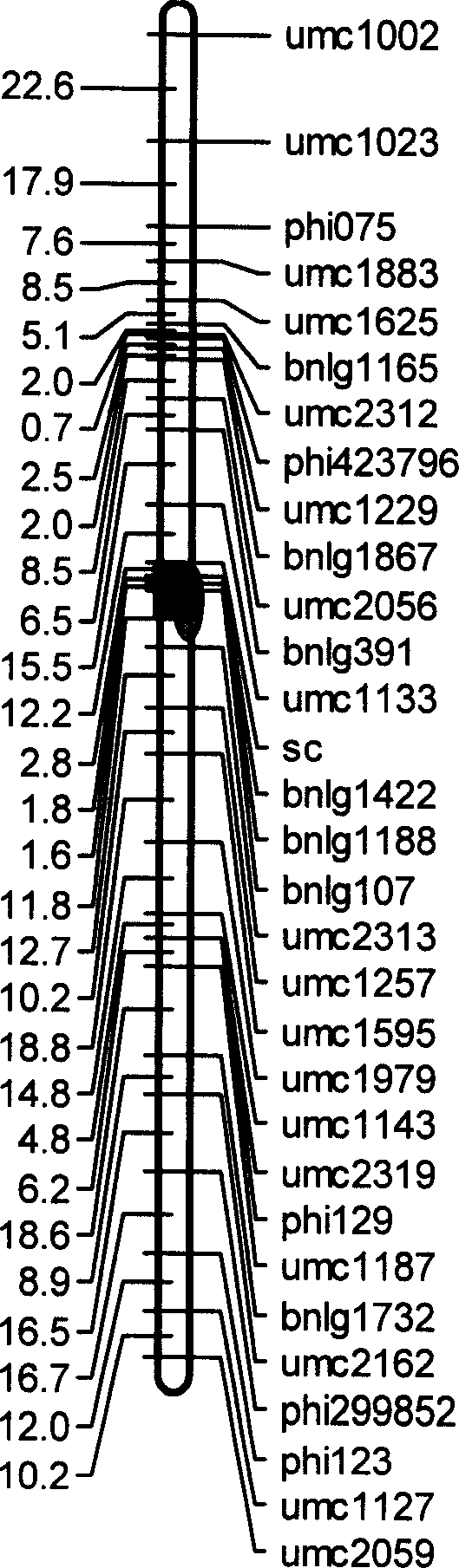
[0030] Embodiment 1, the acquisition of oilc6-m1
[0031] 1. Prepare materials
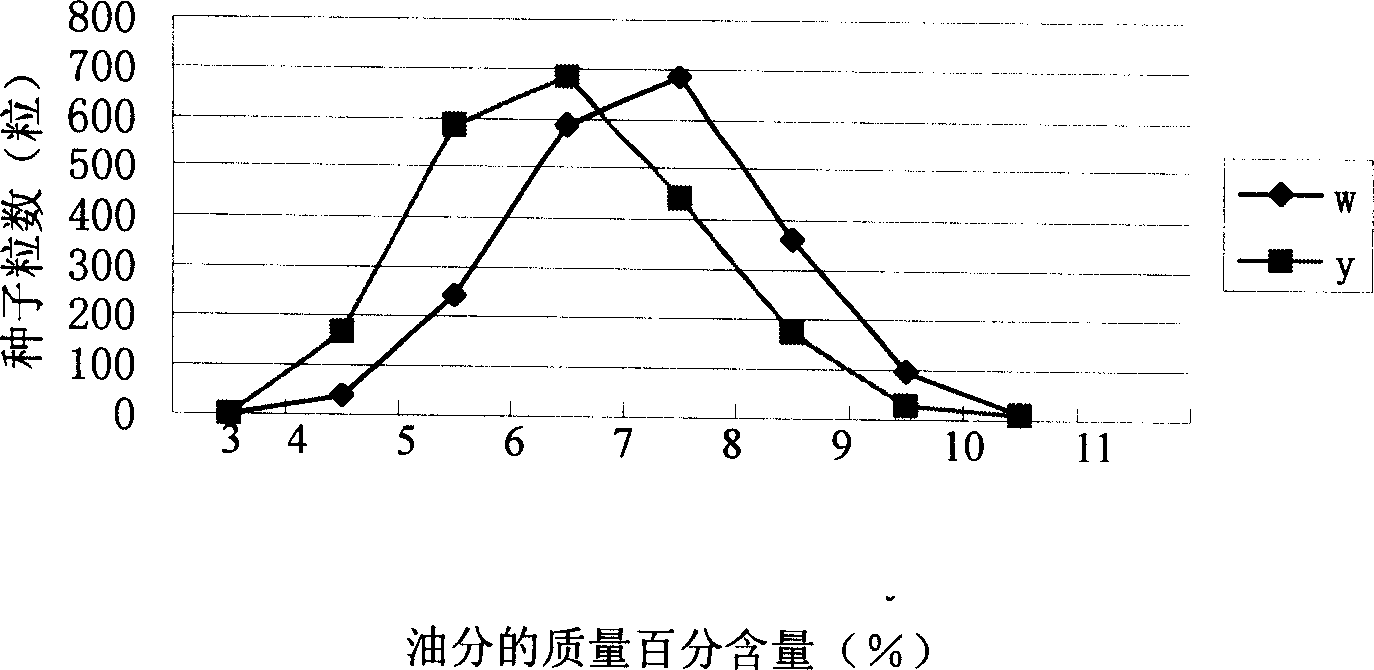
[0032] The high-oil maize mutant CE03005(♀) with purple stems and white kernels was used as the female parent, and the oil content in the kernels was 8.198%. F of 214 individual plants 2 Collect the leaves of a single plant at the big trumpet stage, put them in a -70°C refrigerator for later use, and harvest F 2 The seeds produced by the plant (F 3 grains) to measure the oil content. from F 1 Randomly select 600 seeds from the seeds and plant them on the farm, and harvest self-bred F 2 The seeds produced (F 3 grains) to measure the oil content. Will F 2 The resulting seeds were planted in the experimental station, complete block design, three repetitions, mixed pollination in the plot, and harvested mixed pollinated F 3 The resulting seeds were tested for their oil content. Determination of grain oil content. The results of the frequency distribution of the grain oil content of this gro...
Embodiment 2
[0043] Example 2, using primers p-bnlg1142, and / or p-umc2313, and / or p-bnlg1188, and / or p-bnlg107 to assist in screening high-oil maize varieties
[0044] 1. The experimental materials include the high-oil parent CE03005 (the oil content of the grain is 8.198%) and the F of the high-oil corn mutant selected from the high-oil offspring. 3∶4 (F 3∶4 Refers to F 3 The kernels on the plant are actually F 4 15 high-oil lines (the oil content of the grains are all greater than 6.3%, and the highest is 9%).
[0045] According to the method of Example 1, the following operations are carried out: the genomic DNA of the above-mentioned corn variety is extracted, and the genomic DNA of the above-mentioned corn variety is used as a template, respectively with P-bnlg1422 (sequence 1 and sequence 2 in the sequence table), p-umc2313 (sequence Sequence 3 and sequence 4 in the list), p-bnlg1188 (sequence 5 and sequence 6 in the sequence listing) and p-bnlg107 (sequence 7 and sequence 8 in th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com