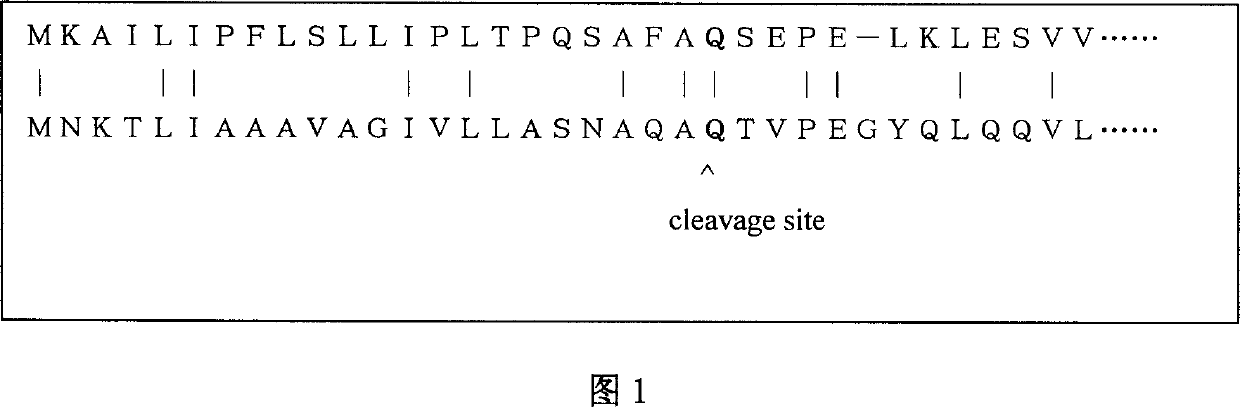
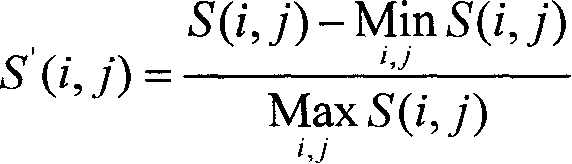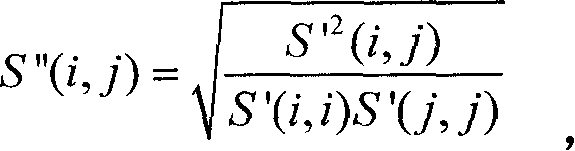Realization of kernel function predicted signal peptide and broken point based on sequence ratio
A technology for sequence alignment and implementation methods, applied in the field of bioengineering, can solve problems such as the low rate of correct judgment of the breakpoint position, lack of physical explanation of the problem, and difficulty in predicting signal peptides.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] The technical solution of the present invention will be further described in detail below in conjunction with specific embodiments.
[0028]The database utilized in the present invention adopts Nielsen (Nielsen, H., Engelbrecht, J., Brunak S., and von Heijne, G. (1997): "Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites" "Protein Eng .”, 1997, 10, pp.1-6). The present invention predicts the Human database, E.coli database, Gram-database and Gram+ database, and the number of signal peptide sequences and non-signal peptide sequences contained in each set of data are 416 and 251, 105 and 119, 266 and 186, 141 respectively and 64. Each amino acid sequence data includes sequence category information, sequence amino acid arrangement and breakpoint position.
[0029] The whole system implementation process is as follows:
[0030] 1. Digitization of attributes.
[0031] Each group of data is processed separately, and the E.c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com