Method for diagnosing non-small cell lung cancers
A non-small cell lung cancer, cell technology, applied in the field of biology, cancer research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0273] [Example 1] General method
[0274](1) Patient and tissue samples
[0275] Following the principle of informed consent, primary lung cancer tissues were obtained from 37 patients (15 women and 22 men, 46-79 years old; average age 66.0) who underwent lobectomy. Obtain clinical information from the medical record, and diagnose each tumor according to histopathological subtype and pathologist grade; 22 of the 37 tumors are classified as adenocarcinoma, 14 are SCC, and the remaining 1 is gland Squamous cell carcinoma. According to the UICC TNM classification standard, the clinical stage of each tumor is judged. All samples were immediately frozen and embedded in TissueTek OCT medium (medium) (Sakura, Tokyo, Japan), and then stored at -80°C.
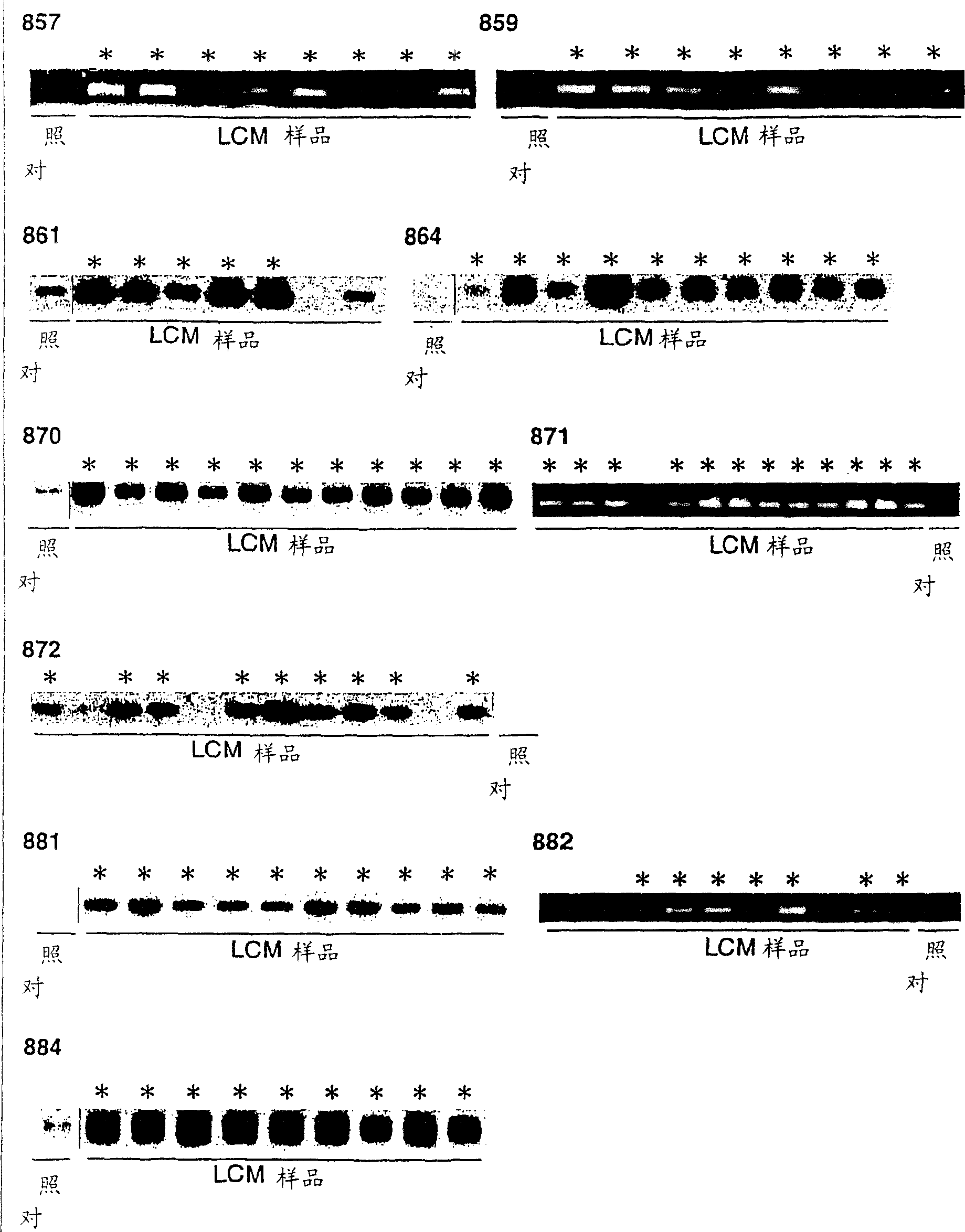
[0276] (2) Laser-capture microdissection (laser-capture microdissection), RNA extraction and base RNA amplification at T7
[0277] Laser-capture microdissection (Kitahara et al., Cancer Res 61: 3544-9 (2001)) was used to selectively coll...
Embodiment 2
[0284] [Example 2] Using clinically relevant expression patterns in non-small cell lung cancer cells to identify genes
[0285] Using the data obtained from the expression profiles of all 37 NSCLC samples, a two-dimensional hierarchical clustering algorithm was applied to analyze the similarity between the samples and genes. When the fluorescence intensity of Cy3- or Cy5- is lower than the cut-off value, such genes are excluded from further analysis, as described above (yanagawa et al., Neoplasia 3:395-401 (2001)), and selected Genes with credible values were obtained in more than 95% of the tested cases. Genes with an observed standard deviation <1.0 are also excluded. The 899 genes that passed through this retention filter paper were further analyzed.
[0286] On the sample axis (horizontal axis), 39 samples from 37 cases (two cases were tested in duplicate to verify the reproducibility and reliability of the experimental method) were divided into 2 main subjects according to ...
Embodiment 3
[0288] [Example 3] Identification and characterization of molecular targets for inhibiting the growth of non-small cell lung cancer cells
[0289] In order to identify and characterize new molecular targets that can regulate the growth, proliferation and survival of cancer cells, antisense S-oligonucleotide technology is used to select target genes as follows.
[0290] (1) Identification of the full-length sequence
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com