Method for identifying fast-growing fish in different salinity
A technology of salt concentration and fish, which is applied in the direction of biochemical equipment and methods, fish farming, microbial measurement/inspection, etc., can solve the problems of inability to identify tilapia strains, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0025] The most basic embodiment of the present invention is a kind of fish that can be selected according to the prolactin genotype of the fish (tilapia is preferred), and then raised and reproduced in a saline environment compatible with its prolactin genotype to obtain the best method of growth.
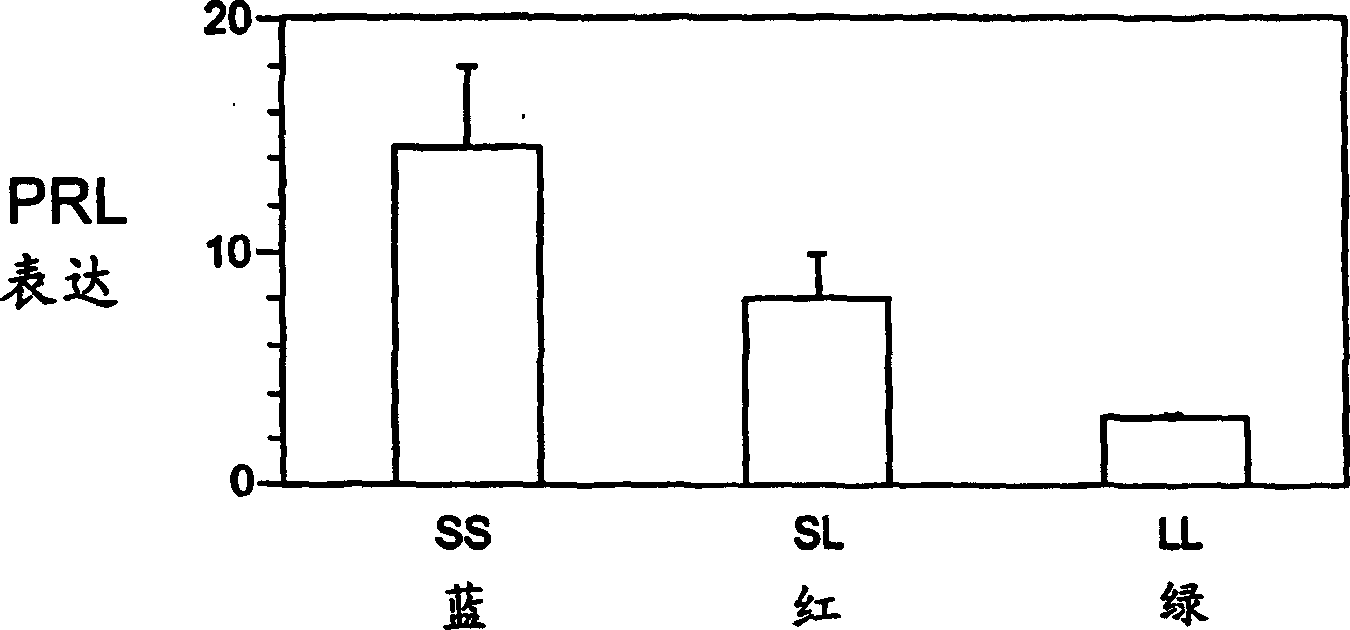
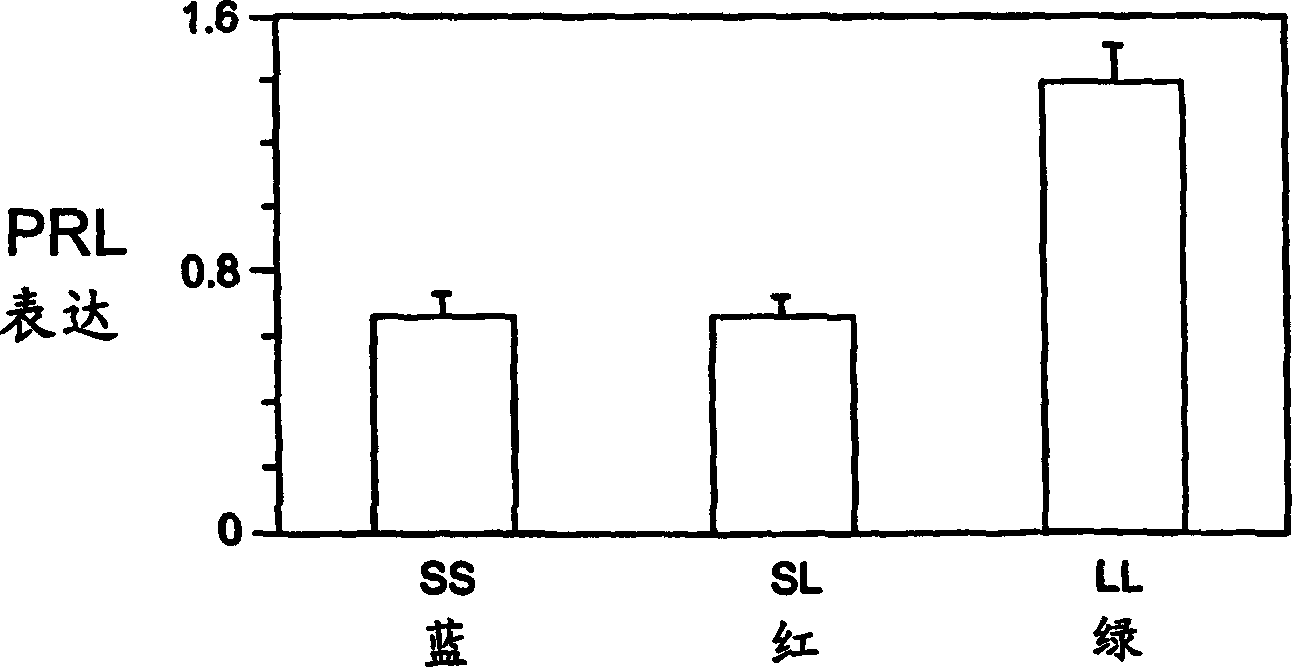
[0026] As noted, past experiments suggested that mRNA levels of prolactin 1 and 2, as well as serum prolactin levels, decreased when tilapia were moved to a saltier environment, with this effect being more pronounced for prolactin 1. Therefore, the applicant's experiments and examples all refer to prolactin 1, but the method of the present invention is not limited to prolactin 1 only.
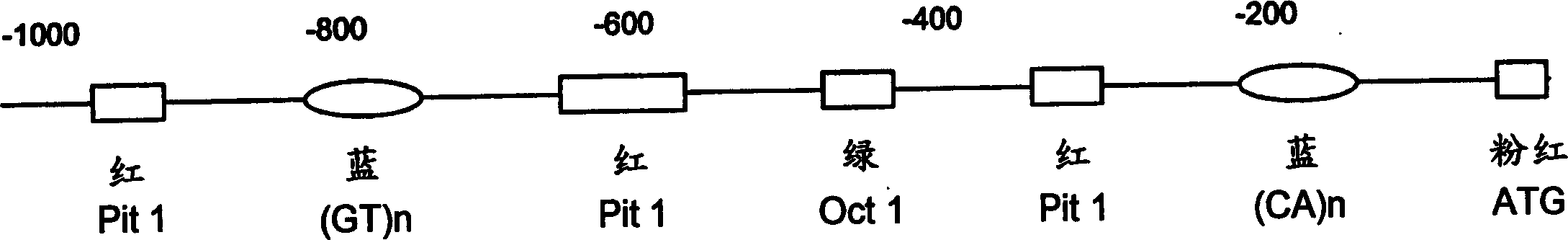
[0027] At the time of development of the present invention, applicants initially focused on the microsatellite closest to the start of the prl1 transcribed region (approximately 200 base pairs bp). Applicants mated Oreochromis (saltwater adapted) females from Mozambique with Nile Oreochromis (fres...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com