Colorectal cancer blood detection marker and application thereof
A technology for colorectal cancer and blood detection, which is applied in the field of tumor detection, can solve the problems of unsatisfactory blood detection results for colorectal cancer, and achieve the effect of improving sensitivity, high specificity, and maintaining specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] 1. Extraction of DNA template:
[0030] When the sample used is a whole blood sample, the blood / cell / tissue genomic DNA extraction kit (catalogue number: DP304) of Tiangen Biochemical Technology (Beijing) Co., Ltd. is used. The volume of the used whole blood sample is 1 ml, and the extracted See the kit instructions for steps.
[0031] When the sample used is a plasma sample, use the nucleic acid extraction reagent of Wuhan Amison Life Science and Technology Co., Ltd. (Ehan Jibei No. 20210740) to extract plasma cfDNA. The volume of plasma used is 600 μL. For specific operations, refer to the kit instruction manual.
[0032] 2. Conversion of sulfite
[0033] The extracted whole blood genomic DNA or plasma cfDNA was subjected to bisulfite conversion, and the nucleic acid conversion kit used was the nucleic acid purification reagent of Wuhan Amison Life Science and Technology Co., Ltd.
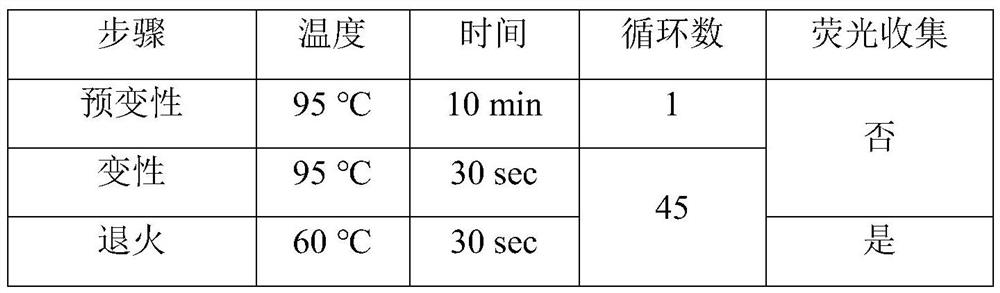
[0034] 3. Methylation-specific PCR reaction
[0035] Perform methylation-specific P...
experiment example 1
[0049] 30 plasma samples from patients with advanced colorectal adenoma, 50 plasma samples from patients with colorectal cancer, and 30 plasma samples from healthy people from blood donation were collected from Zhongnan Hospital of Wuhan University. All patients were anonymized. According to the method described in Example 1, the plasma sample DNA was extracted, the extracted DNA was subjected to bisulfite conversion, and the converted DNA was used as a template to perform a methylation-specific PCR reaction, and the ITGA4 gene, ZNF829 gene and BEND5 gene were tested. The sensitivity of these three genes in adenoma and cancer samples and the specificity in healthy samples were calculated for the methylation levels in various plasma samples, and the results are shown in Table 3.
[0050] Table 3 Sensitivity and specificity data of three genes in adenoma, cancer and healthy plasma
[0051]
[0052] It can be seen from Table 3 that the sensitivity of methylation of the three g...
experiment example 2
[0054] 60 cases of whole blood samples from healthy people (different from the healthy people in Experimental Example 1) were collected from Zhongnan Hospital of Wuhan University, and all patients were anonymized. According to the method described in Example 1, the whole blood sample DNA was extracted, the extracted DNA was converted to bisulfite, and the converted DNA was used as a template to perform a methylation-specific PCR reaction, and the ITGA4 gene, ZNF829 gene and BEND5 were tested. For the methylation levels of the genes in 60 samples, the methylation positive rates of the three genes in the healthy samples were counted respectively, and the specificity was calculated. The results are shown in Table 4.
[0055] Table 4 The specificity of three genes in 60 cases of healthy people whole blood samples
[0056] gene name positive number specificity ITGA4 5 91.67% ZNF829 12 80.00% BEND5 6 90.00%
[0057] As shown in Table 4, the spe...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com