Exonuclease III-driven three-dimensional DNA nanomachines and their applications
A nanomachine and exonuclease technology, applied in the field of biological analysis, can solve the problems of poor sensitivity, complicated operation and high background signal, and achieve the effect of high sensitivity and improved detection sensitivity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
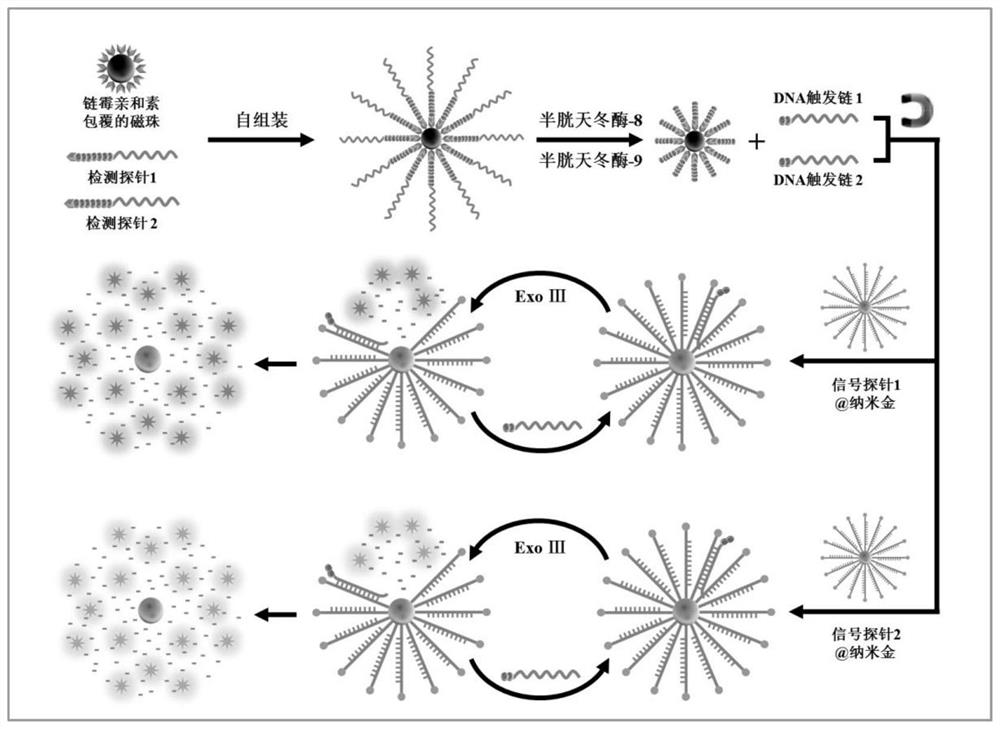
[0061] Example 1 Construction of exonuclease III-driven three-dimensional DNA nanomachines
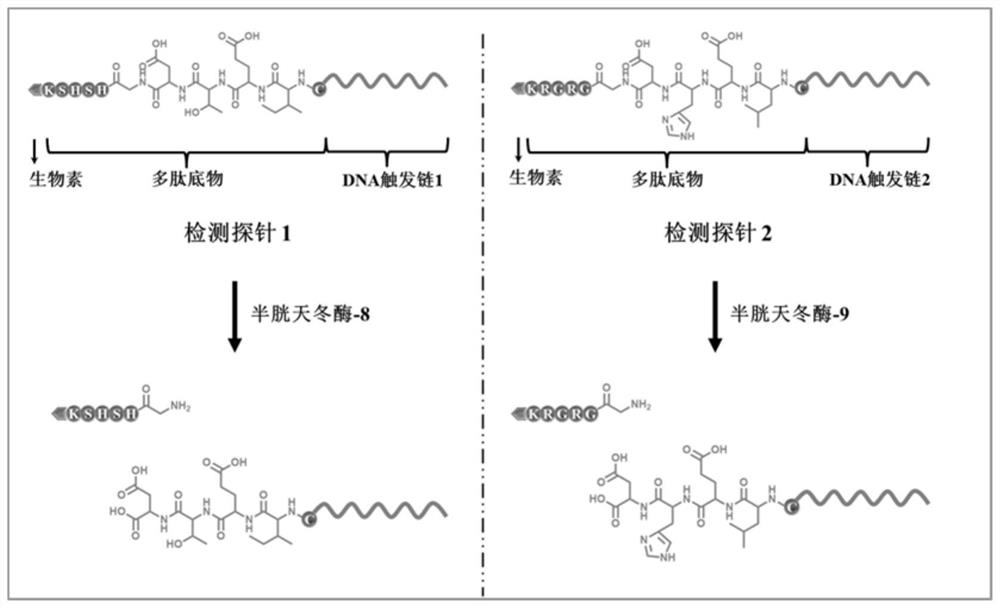
[0062] The detection probe is a polypeptide-DNA detection probe, which includes two kinds. The polypeptide chain of the detection probe 1 contains the recognition site Ile-Glu-Thr-Asp (IETD) of caspase-8, and its DNA part It can be complementary to signal probe 1; the polypeptide chain of detection probe 2 contains the recognition site Leu-Glu-His-Asp (LEHD) of caspase-9, and its DNA part can be complementary to signal probe 2 .
[0063] The sequence information involved in the exemplary embodiment of the present invention is shown in Table 1:
[0064] Table 1
[0065]
[0066]
[0067] Note: The recognition site of the target in the detection probe is shown in bold. The detection probe is a polypeptide-DNA structure, wherein the amino terminus of the polypeptide is modified on the 5' end of the DNA sequence (the structure of the carboxyl and amino groups in the probe struct...
Embodiment 2
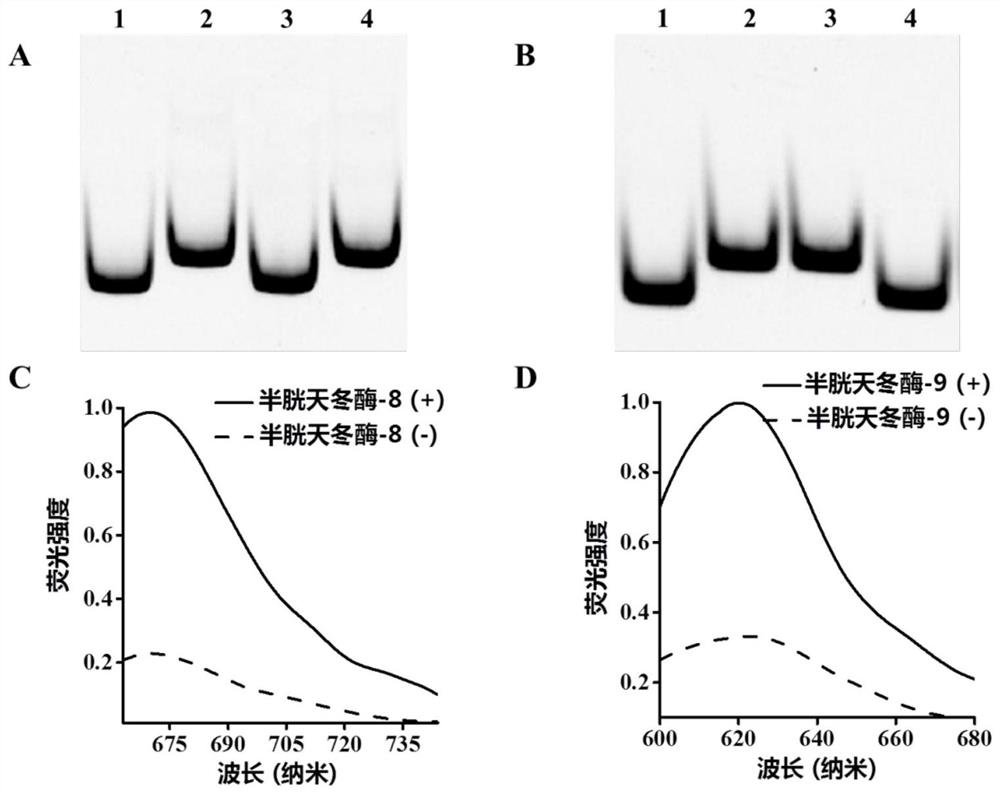
[0071] Example 2 Detection of caspase activity and detection of caspase under activity inhibition
[0072] 1. Detection of caspase activity: The step of detection of caspase activity includes two steps, which are the cleavage of the detection probe by caspase and the fluorescence caused by the three-dimensional DNA nanomachine driven by exonuclease III. Cyclic release of molecules. The cleavage of detection probes by caspase was carried out in a 20 μl reaction system containing 11.5 μl of 1× phosphate buffer solution, 8 μl of detection probe-magnetic beads and 0.5 units of Caspase-8 or / and Caspase-9, incubate for 1 hour at 37°C. After magnetic separation, the supernatant containing the cleavage product was taken for the next reaction. The reaction system of the exonuclease III-driven three-dimensional DNA walker in the second step is 20 μl, which contains 1× Cutsmart buffer (500 mmol / L potassium acetate, 200 mmol / L trihydroxyaminomethane-acetic acid) , 100 mmol / L magnesiu...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com