SCAR marker and method for early identification of tomato internode length
A tomato and internode technology, applied in the fields of biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of difficult new varieties, low efficiency, and few molecular markers, achieving high accuracy and speeding up selection. The effect of low-cost growth and detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Obtaining of embodiment 1 SCAR mark
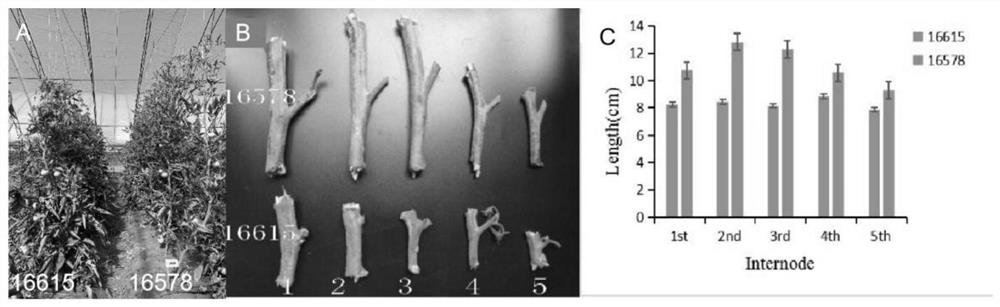
[0018] Using wild-type tomato 16615 and long-node field natural mutant 16578 as materials, construct P 1 ,P 2 , F 1 and F 2 Four-generation population, using parental resequencing combined with F 2 The group separation method (Bulked Segregant Analysis, BSA) further screened candidate intervals. The specific operation is as follows: select the parent parent P 1 ,P 2 One each, F 2 25 strains in each extreme mixed pool were used for DNA extraction; library construction and sequencing were carried out, and the whole genome resequencing on the BGISEQ platform was completed by Shenzhen Huada Company; further biological information analysis was performed, and the filtered reads were compared with the reference genome. NCBI: https: / / www.ncbi.nlm.nih.gov / genome / gdv / browser / genome / ? id=GCF_000188115.4 as a reference genome. Through bioinformatics analysis, unqualified data is filtered out to obtain high-quality valid data. Finally,...
Embodiment 2
[0026] Application of embodiment 2THH-FOR0 mark
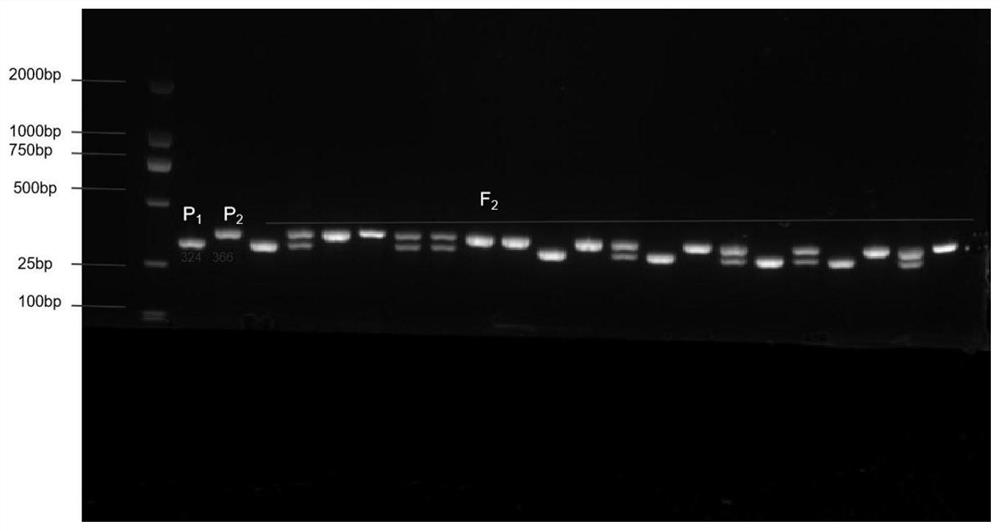
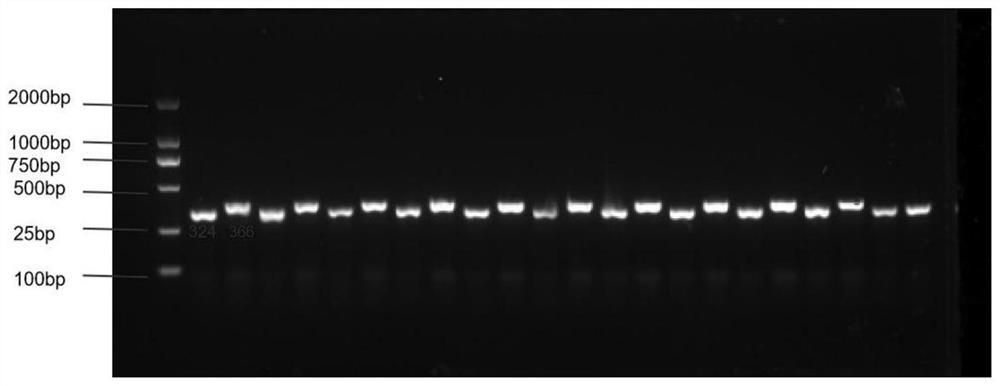
[0027] F 1 , F 1 F 2 group. Using the SCAR marker primer set for 210 copies of F 2 Segregate populations for internode length identification. The specific method is as follows: first, the CTAB method is used to extract leaf DNA of tomato with identification; the leaf DNA is used as a template, and the primer pair in the embodiment is used for amplification. The reaction system for PCR amplification is: 10 μL 2×Mix Master, 1 μL DNA template, 1 μL each of upstream and downstream primers, 7 μL ddH 2O. The reaction program of PCR amplification is: pre-denaturation at 94°C for 5 min; denaturation at 94°C for 30 s; annealing at 56°C for 40 s; extension at 72°C for 50 s; cycle 35 times; extension at 72°C for 10 min; . The amplified products are then identified.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com