CD81 aptamer and application thereof
An aptamer and nucleic acid aptamer technology, applied in the biological field, can solve the problems of exosome purity, integrity and process maturity, high price, and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Example 1. Screening of aptamers targeting CD81
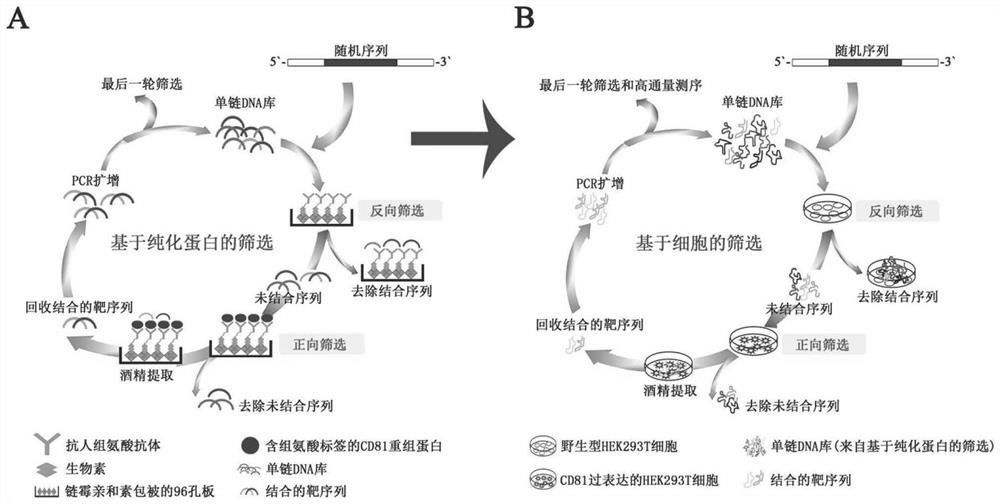
[0072] The aptamer screening of CD81 is based on the SELEX technology of CD81 recombinant protein and CD81 positive expression cell line (the schematic diagram is shown in figure 1 shown), the specific way is: in about 10 14 Aptamers targeting CD81 were screened within a library of aptamer-ssDNA. The DNA in the library all contains the following core sequence (86nt, wherein N is any other base): 5'-TAG GGA AGAGAA GGA CAT ATG AT-40N-TTG ACT AGT ACA TGA CCA CTT GA-3'. The library contains 671 DNA sequences shown in Table 12. Aptamers bound to CD81 recombinant protein or CD81-positive cell lines were eluted and amplified by PCR. The sequences of the upstream and downstream primers for PCR amplification are: FITC-5'-TA GGG AAG AGAAGG ACA TAT GAT-3' and 5'-TTT TTT TTT TTT TTT TTT TT / iSp9 / T CAA GTG GTC ATG TACTAG TCA A- 3'. The amplified DNA was identified by high-throughput sequencing, and 6 aptamers with the best bindin...
Embodiment 2
[0075] Example 2. Identification of binding ability of CD81 aptamer and CD81 at cell level
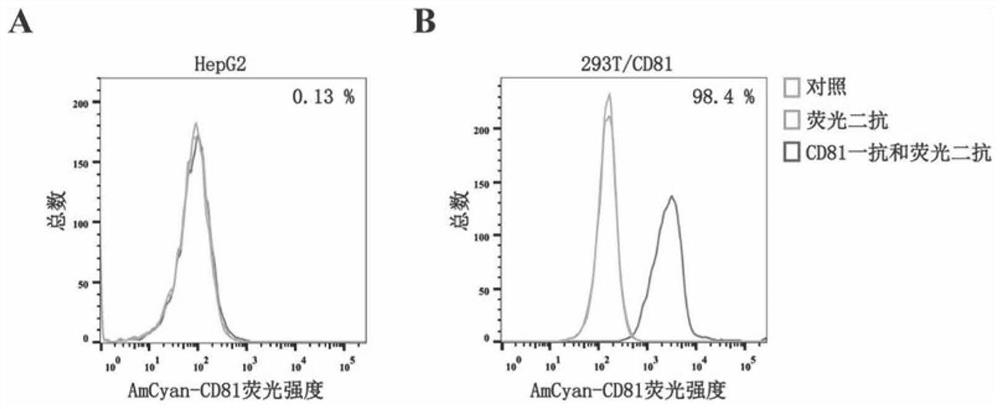
[0076] 1. Construction of recombinant animal cells overexpressing CD81
[0077] The specific construction method of the recombinant animal cell (the cell is named 293T / CD81) overexpressed CD81 is as follows: the KpnI and XbaI of the plasmid pCMV3-C-His (Beijing Yiqiao Shenzhou Technology Co., Ltd., the product of Sino Biological Inc.) The sequence between the recognition sites is replaced by the cDNA sequence of human CD81, and the recombinant expression vector that keeps other parts of the pCMV3-C-His sequence unchanged, the recombinant expression vector is named pCMV3-CD81. pCMV3-CD81 was introduced into HEK293T cells to obtain recombinant cells, which were named 293T / CD81. The nucleotide sequence of the cDNA of human CD81 is the nucleotide sequence shown in 7 in the sequence table, and its encoded amino acid sequence is the protein with his tag shown in the sequence.
[0078] 2. T...
Embodiment 3
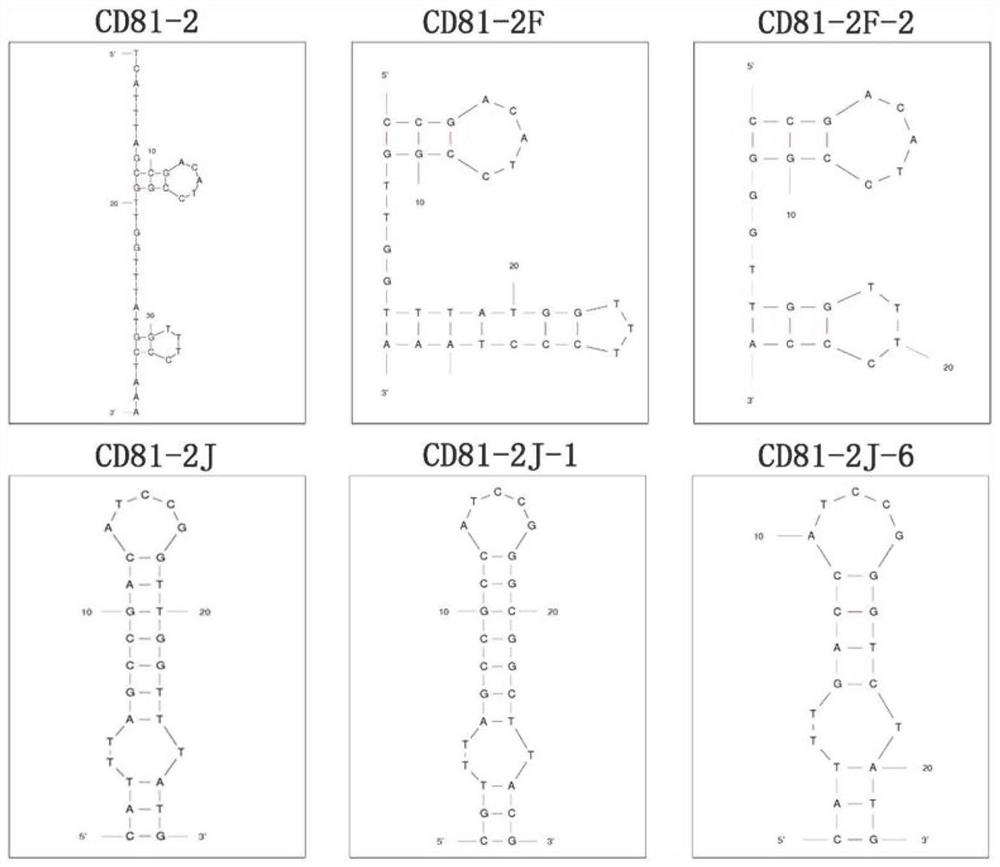
[0090] Example 3. Identification of binding ability of CD81-2J and CD81-2F subtypes
[0091] For CD81-2J and CD81-2F, we constructed 8 subclones respectively, and the sequences are shown in Table 5 and Table 6. By the test method in Example 2, compare the binding force of the subtypes of CD81-2J and CD81-2F as follows: Figure 8-11 shown. The results demonstrated that subtypes CD81-2J-1, CD81-2J-6 and CD81-2F-2 had significantly higher binding ability to CD81-overexpressing HEK293T cells (293T / CD81) than negative control HepG2 cells (without CD81) (* *p<0.1, ***p<0.01), indicating that the above three subtypes have stronger binding ability to CD81 in its natural conformation. Therefore, CD81-2J-1, CD81-2J-6 and CD81-2F-2 were selected for subsequent verification and detection experiments.
[0092] Table 5 Sequences of 8 subtypes of aptamer CD81-2J
[0093]
[0094]
[0095] Table 6 Sequences of 8 subtypes of aptamer CD81-2J
[0096]
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle size | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com