Probe design method and positioning method for wheat exon sequencing gene positioning
A technology for exon sequencing and gene mapping, which is applied in the fields of biochemical equipment and methods, genomics, and microbial determination/inspection, and can solve the problem of high cost of genetic information.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
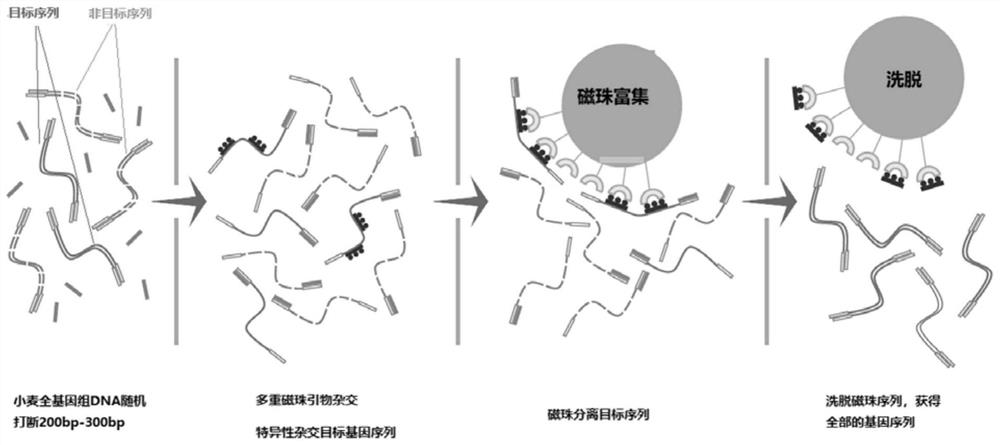
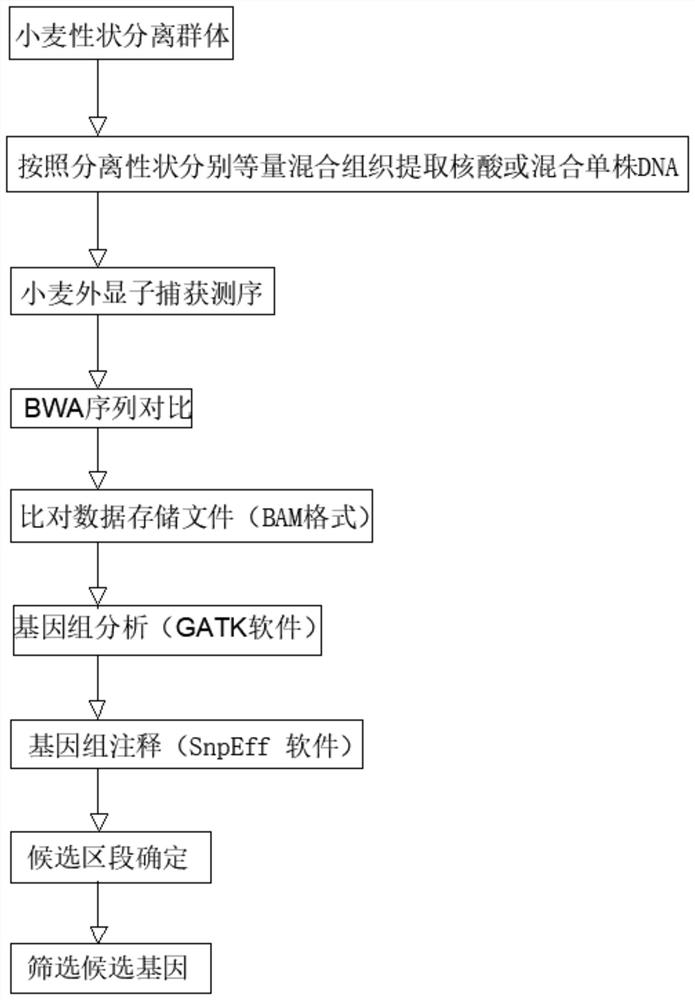
[0034] A probe design method for wheat exome sequencing gene localization, comprising the steps of:
[0035] Step 1. Select different tissues, environments, and developmental data through large-scale transcriptome sequencing data, and then perform sequencing on the second-generation Illumina high-throughput sequencing platform to obtain sequencing data for use;
[0036] Step 2: Use the STAR software to compare and analyze the sequencing data and the comparison reference genome, and then use the stringtie function to splice the transcripts, and filter the transcripts whose gene expression level TPM is less than 2, and keep the transcripts with TPM ≥ 2 for later use; The comparison reference genome is specifically IWGSCRefSeq (https: / / www.wheatgenome.org / );
[0037] Step 3. Merge the transcript in step 2 with the IWGSC CS Annotation transcript to obtain the merged transcript for use;
[0038] Step 4. Use TransDecoder (v5.5.0) to perform ORF prediction on the merged transcripts ...
Embodiment 2
[0053] A probe design method for wheat exome sequencing gene localization, comprising the steps of:
[0054] Step 1. Select different tissues, environments, and developmental data through large-scale transcriptome sequencing data, and then perform sequencing through the BGI BIGseq high-throughput sequencing platform to obtain sequencing data for use;
[0055] Step 2: Use the STAR software to compare and analyze the sequencing data and the comparison reference genome, and then use the stringtie function to splice the transcripts, and filter the transcripts whose gene expression level TPM is less than 2, and keep the transcripts with TPM ≥ 2 for later use; The comparison reference genome is specifically IWGSC RefSeq (https: / / www.wheatgenome.org / );
[0056] Step 3. Merge the transcript in step 2 with the IWGSC CS Annotation transcript to obtain the merged transcript for use;
[0057] Step 4. Use TransDecoder (v5.5.0) to perform ORF prediction on the merged transcripts in step 3,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com