Method for identifying transgenic soybean plants through flower color
A technology of transgenic plants and flower colors, applied in botany equipment and methods, genetic engineering, plant genetic improvement, etc., can solve the problems of destroying tissue activity and not being easy to observe
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
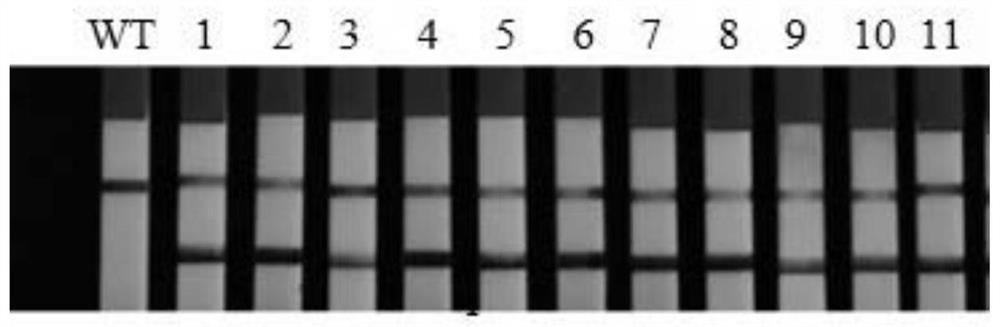
Image
Examples
Embodiment 1
[0050] Embodiment 1. A method for identifying transgenic soybean plants by flower color
[0051] 1. Cloning of GmZI gene
[0052] RNA was extracted from the flower of the purple-flowered soybean cultivar Zhonghuang42, and cDNA was synthesized by reverse transcription.
[0053] The cDNA was used as a template, and the primer pair composed of the new primer F and primer R was used for PCR amplification, and the product size was about 1500bp.
[0054] F: 5'-ATGGACTCATTGTTACTTCT-3';
[0055] R: 5'-CAACCAATTCTAAGAAATGTAA-3'.
[0056] After sequencing, the nucleotide sequence of the PCR product is sequence 1 in the sequence table, which is the ZI gene.
[0057] Two, the construction of GmZI overexpression vector
[0058] 1. The PCR product obtained in the above one is used as a template, and a primer pair consisting of primer F-inf and primer R-inf is used for PCR amplification, and a PCR amplification product of about 1500 bp is recovered.
[0059] F-inf: 5'-GAGAACACGGGGGACTCT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com