Crispr double nickase based amplification compositions, systems, and methods
A technology of nicking enzymes and complexes, applied in biochemical equipment and methods, hydrolytic enzymes, genetic engineering, etc., can solve problems such as application limitations and low sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0331] Example 1 - CRISPR-Nickase-Based Amplification (CRISPR-NEAR) and NEAR SHERLOCK Detection
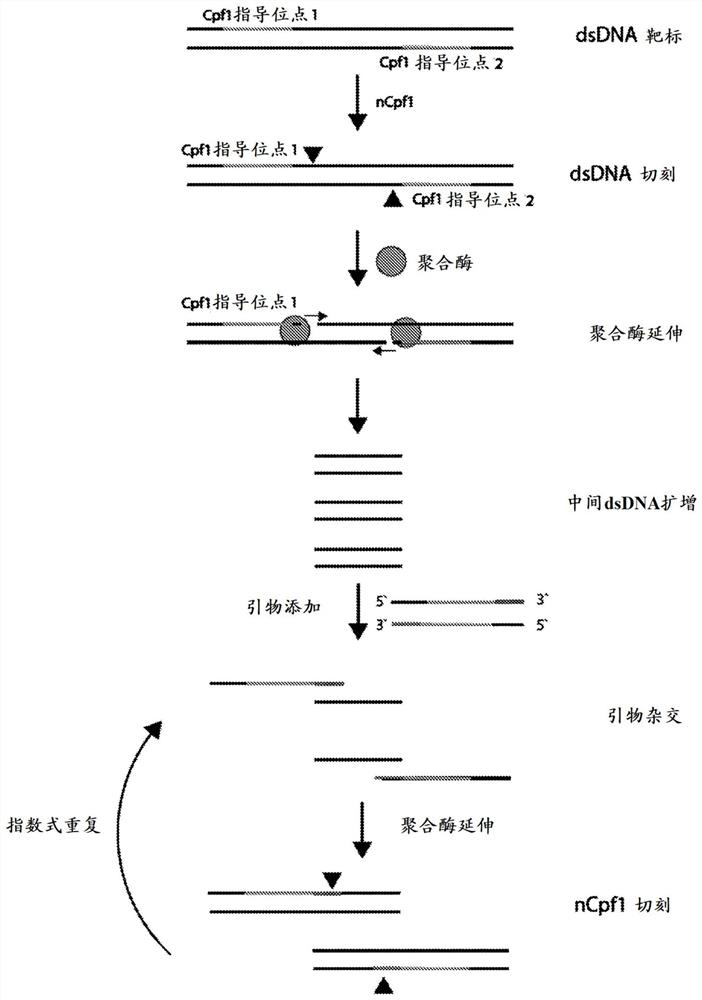
[0332] In this example, nicking enzyme-based amplification was tested using a CRISPR-Cas enzyme called CRISPR-NEAR combined with a CRISPR SHERLOCK detection method. figure 1 A schematic diagram of nickase-based amplification using CRISPR-Cas enzymes is shown.
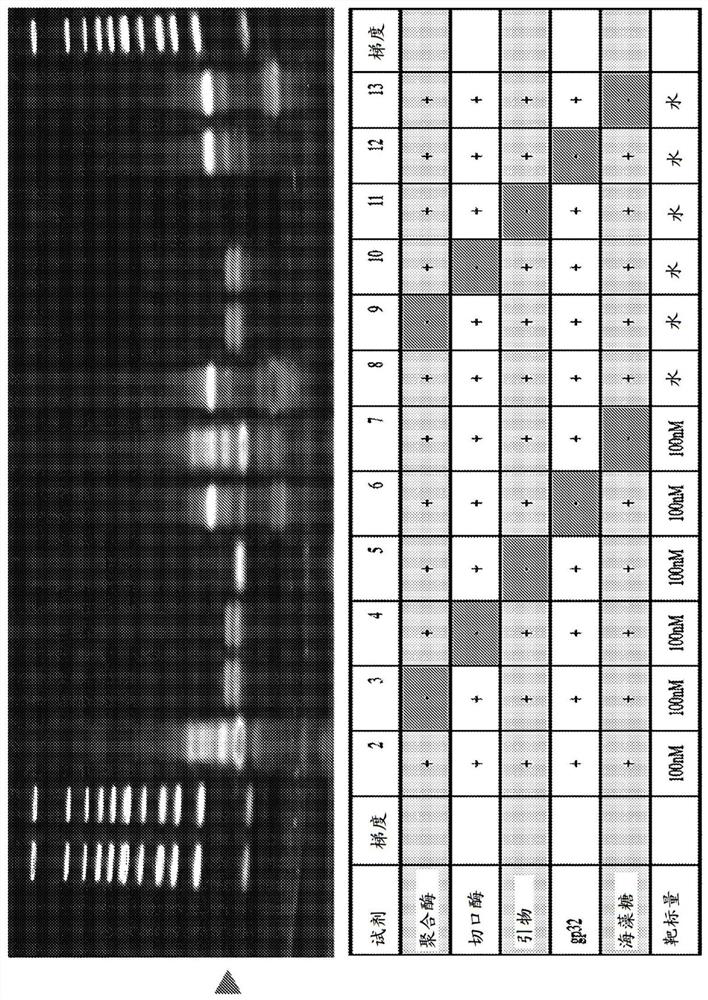
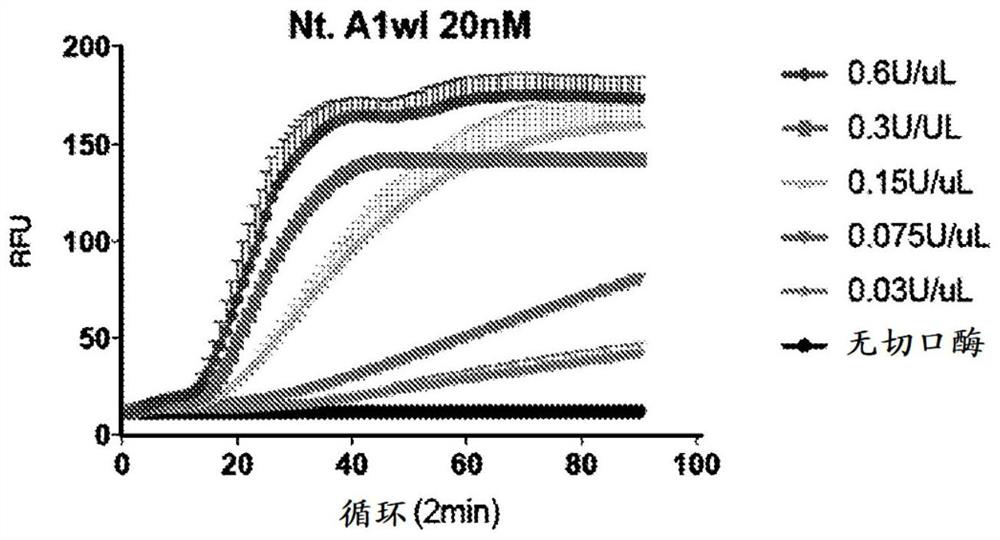
[0333] CRISPR-NEAR can be performed using DNA or RNA input. CRISPR-NEAR is also compatible with downstream SHERLOCK assays by incorporating T7 promoter sequences in the amplification primers. Figure 9 A schematic diagram of the combination of CRISPR-NEAR with the SHERLOCK assay is shown. One of the main advantages of using CRISPR-NEAR is that it can be amplified much faster than RPA. The method uses very simple buffers that allow easy combination of all steps of the SHERLOCK assay into one reaction. On the other hand, RPA amplification uses very viscous buffers that are difficult to use with other reagents.
[0334] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com