Fusion gene nucleic acid detection method based on combination of capillary electrophoresis fragment analysis and first-generation sequencing
A technology of capillary electrophoresis and gene fusion, applied in biochemical equipment and methods, measurement/testing of microorganisms, DNA/RNA fragments, etc., can solve problems such as inability to obtain sequence information and cumbersome operation steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Example 1. Fusion gene nucleic acid detection method based on the combination of capillary electrophoresis fragment analysis and first-generation sequencing 1. Design of multiple amplification primers
[0069] According to the connection sequence of multiple target fusion genes (the sequence containing the connection site of the fusion gene is 100-300bp in size) as the target fragment, set multiple pairs of amplification primers for multiple nucleic acid detection, and each pair of amplification primers is specific Amplify 1 target fusion gene:
[0070] Each pair of amplification primers consists of primer A and primer B, wherein:
[0071] Starting from the 5' end, the primer A is sequentially composed of an adapter sequence A for binding the sequencing primer and a specific sequence A specifically binding to the target fragment, and the 5' end of the primer A is fluorescently modified. Among them, the fluorescently modified groups can be, for example, FAM, HEX, VIC, T...
Embodiment 2
[0102] Example 2. Application of fusion gene nucleic acid detection method based on the combination of capillary electrophoresis fragment analysis and generation sequencing
[0103] 1. Design of Multiplex Amplification Primers
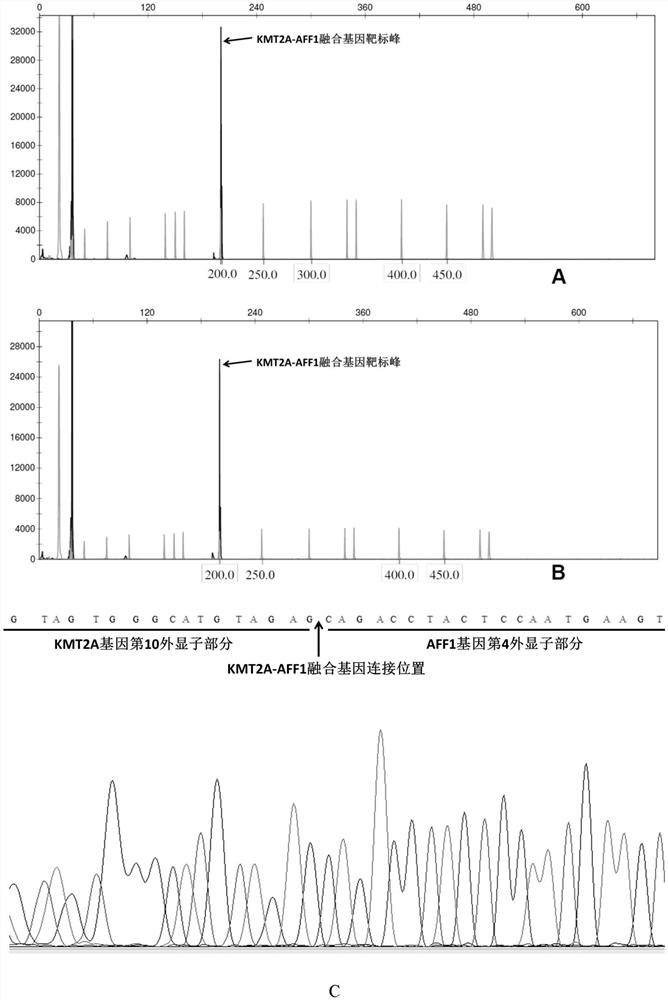
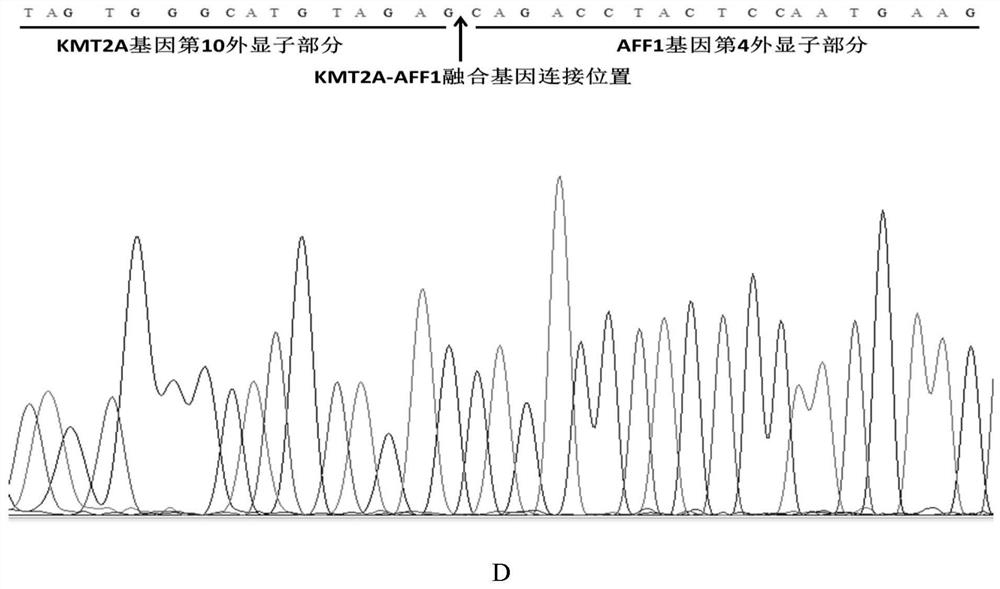
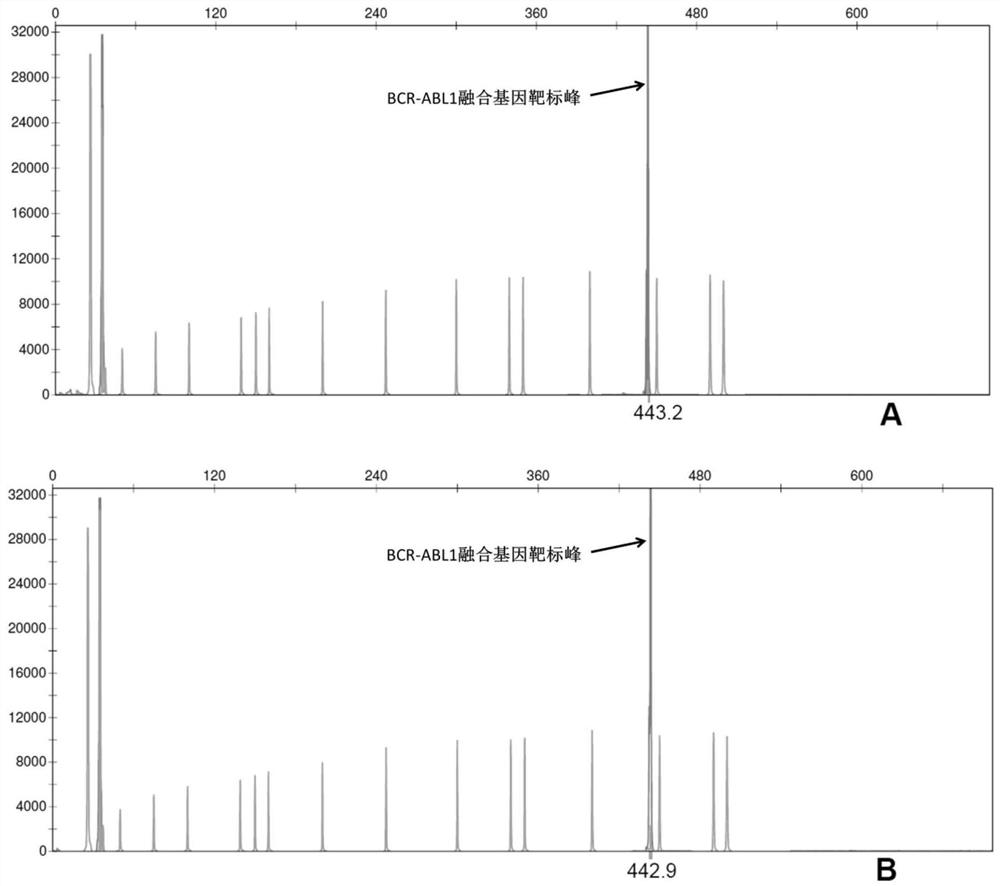
[0104] Amplification primer pairs were designed for the four common fusion genes KMT2A-AFF1, BCR-ABL1, CBFB-MYH11 and TCF3-PBX1 in leukemia.
[0105] Select the junction sequence of each fusion gene (the size of the junction site containing the fusion gene is within the sequence of 200bp) as the target fragment, and set up multiple pairs of amplification primer pairs (shown in Table 1) for multiple nucleic acid detection respectively. The primer pair specifically amplifies a target fusion gene:
[0106] Among them, the upstream primer of KMT2A-AFF1 is marked with FAM (the corresponding color is blue), the downstream primer of BCR-ABL1 is marked with HEX (the corresponding color is green), and the upstream primer of CBFB-MYH11 is marked with TAMRA (the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com